Phenotypic heterogeneity driven by plasticity of the intermediate EMT state governs disease progression and metastasis in breast cancer
- PMID: 35921406
- PMCID: PMC9348802
- DOI: 10.1126/sciadv.abj8002
Phenotypic heterogeneity driven by plasticity of the intermediate EMT state governs disease progression and metastasis in breast cancer
Abstract
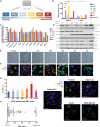
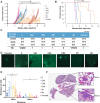
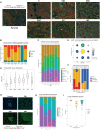
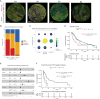
The epithelial-to-mesenchymal transition (EMT) is frequently co-opted by cancer cells to enhance migratory and invasive cell traits. It is a key contributor to heterogeneity, chemoresistance, and metastasis in many carcinoma types, where the intermediate EMT state plays a critical tumor-initiating role. We isolate multiple distinct single-cell clones from the SUM149PT human breast cell line spanning the EMT spectrum having diverse migratory, tumor-initiating, and metastatic qualities, including three unique intermediates. Using a multiomics approach, we identify CBFβ as a key regulator of metastatic ability in the intermediate state. To quantify epithelial-mesenchymal heterogeneity within tumors, we develop an advanced multiplexed immunostaining approach using SUM149-derived orthotopic tumors and find that the EMT state and epithelial-mesenchymal heterogeneity are predictive of overall survival in a cohort of stage III breast cancer. Our model reveals previously unidentified insights into the complex EMT spectrum and its regulatory networks, as well as the contributions of epithelial-mesenchymal plasticity (EMP) in tumor heterogeneity in breast cancer.
Figures


Similar articles
-
Quantifying epithelial-mesenchymal heterogeneity and EMT scoring in tumor samples via tyramide signal amplification (TSA).Methods Cell Biol. 2022;171:149-161. doi: 10.1016/bs.mcb.2022.06.003. Epub 2022 Jul 26. Methods Cell Biol. 2022. PMID: 35953198
-
EMT, cell plasticity and metastasis.Cancer Metastasis Rev. 2016 Dec;35(4):645-654. doi: 10.1007/s10555-016-9648-7. Cancer Metastasis Rev. 2016. PMID: 27878502 Review.
-
Mesenchymal-epithelial transition in lymph node metastases of oral squamous cell carcinoma is accompanied by ZEB1 expression.J Transl Med. 2023 Apr 19;21(1):267. doi: 10.1186/s12967-023-04102-w. J Transl Med. 2023. PMID: 37076857 Free PMC article.
-
Genome-wide CRISPR screen identifies PRC2 and KMT2D-COMPASS as regulators of distinct EMT trajectories that contribute differentially to metastasis.Nat Cell Biol. 2022 Apr;24(4):554-564. doi: 10.1038/s41556-022-00877-0. Epub 2022 Apr 11. Nat Cell Biol. 2022. PMID: 35411083 Free PMC article.
-
Epithelial to mesenchymal transition (EMT) in metaplastic breast cancer and phyllodes breast tumors.Med Oncol. 2023 Dec 16;41(1):20. doi: 10.1007/s12032-023-02259-4. Med Oncol. 2023. PMID: 38104042 Review.
Cited by
-
"In medio stat virtus": Insights into hybrid E/M phenotype attitudes.Front Cell Dev Biol. 2022 Nov 18;10:1038841. doi: 10.3389/fcell.2022.1038841. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36467417 Free PMC article. Review.
-
Metabolic dependencies of metastasis-initiating cells in female breast cancer.Nat Commun. 2023 Nov 4;14(1):7076. doi: 10.1038/s41467-023-42748-8. Nat Commun. 2023. PMID: 37925484 Free PMC article.
-
Dynamics of Epithelial-Mesenchymal Plasticity: What Have Single-Cell Investigations Elucidated So Far?ACS Omega. 2023 Mar 22;8(13):11665-11673. doi: 10.1021/acsomega.2c07989. eCollection 2023 Apr 4. ACS Omega. 2023. PMID: 37033874 Free PMC article. Review.
-
Epithelial-to-Mesenchymal Transition Gene Signature in Circulating Melanoma Cells: Biological and Clinical Relevance.Int J Mol Sci. 2023 Jul 22;24(14):11792. doi: 10.3390/ijms241411792. Int J Mol Sci. 2023. PMID: 37511550 Free PMC article.
-
Increased prevalence of hybrid epithelial/mesenchymal state and enhanced phenotypic heterogeneity in basal breast cancer.iScience. 2024 May 27;27(7):110116. doi: 10.1016/j.isci.2024.110116. eCollection 2024 Jul 19. iScience. 2024. PMID: 38974967 Free PMC article.
References
-
- Thiery J. P., Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442–454 (2002). - PubMed
-
- Ognjenovic N. B., Bagheri M., Mohamed G. A., Xu K., Chen Y., Saleem M. A. M., Brown M. S., Nagaraj S. H., Muller K. E., Gerber S. A., Christensen B. C., Pattabiraman D. R., Limiting self-renewal of the basal compartment by pka activation induces differentiation and alters the evolution of mammary tumors. Dev. Cell 55, 544–557.e6 (2020). - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

