Variability among the Isolates of Broad Bean Mottle Virus and Encapsidation of Host RNAs
- PMID: 35890061
- PMCID: PMC9321246
- DOI: 10.3390/pathogens11070817
Variability among the Isolates of Broad Bean Mottle Virus and Encapsidation of Host RNAs
Abstract
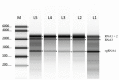
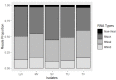
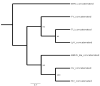
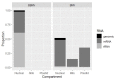
Broad bean mottle bromovirus infects legume plants and is transmissible by insects. Several broad bean mottle virus (BBMV) isolates have been identified, including one in England (isolate Ba) and five in the Mediterranean countries: Libya (LyV), Morocco (MV), Syria (SV), Sudan (TU) and Tunisia (TV). Previously, we analyzed the nucleotide sequence of the Ba RNA and here we report on and compare it with another five Mediterranean variants. The RNA segments in the latter ones were extensively homologous, with some SNPs, single nucleotide deletions and insertions, while the number of mutations was higher in isolate Ba. Both the 5' and 3' untranslated terminal regions (UTRs) among the corresponding RNAs are highly conserved, reflecting their functionality in virus replication. The AUG initiation codons are within suboptimal contexts, possibly to adjust/regulate translation. The proteins 1a, 2a, 3a and coat protein (CP) are almost identical among the five isolates, but in Ba they have more amino acid (aa) substitutions. Phylogenetic analysis revealed the isolates from Morocco and Syria clustering with the isolate from England, while the variants from Libya, Tunisia and Sudan created a different clade. The BBMV isolates encapsidate a high content of host (ribosomal and messenger) RNAs. Our studies present BBMV as a useful model for bromoviruses infecting legumes.
Keywords: RNA genome variability; bromoviruses; cellular RNA encapsidation; phylogeny; plant RNA viruses.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
De novo generation of defective interfering-like RNAs in broad bean mottle bromovirus.Virology. 1995 Oct 1;212(2):574-86. doi: 10.1006/viro.1995.1515. Virology. 1995. PMID: 7571427
-
The nucleotide sequence and genome organization of the RNA-1 segment in two bromoviruses: broad bean mottle virus and cowpea chlorotic mottle virus.Virology. 1991 Dec;185(2):553-62. doi: 10.1016/0042-6822(91)90525-g. Virology. 1991. PMID: 1962437
-
The nucleotide sequence and genome organization of the RNA2 and RNA3 segments in broad bean mottle virus.Virology. 1992 Apr;187(2):671-81. doi: 10.1016/0042-6822(92)90470-a. Virology. 1992. PMID: 1546462
-
Use of a defective RNA of broad bean mottle bromovirus for stable gene expression in legumes.Arch Virol. 2008;153(9):1755-8. doi: 10.1007/s00705-008-0174-y. Epub 2008 Aug 5. Arch Virol. 2008. PMID: 18679766
-
Infectious transcripts from PCR-amplified broad bean mottle bromovirus cDNA clones and variable nature of leader regions in RNA 3 segment.J Gen Virol. 1994 Mar;75 ( Pt 3):693-7. doi: 10.1099/0022-1317-75-3-693. J Gen Virol. 1994. PMID: 8126469
Cited by
-
Characterization of Variant RNAs Encapsidated during Bromovirus Infection by High-Throughput Sequencing.Pathogens. 2024 Jan 22;13(1):96. doi: 10.3390/pathogens13010096. Pathogens. 2024. PMID: 38276169 Free PMC article.
References
-
- Fortass M., Bos L. Broad bean mottle virus in Morocco; variability, interaction with food legume species, and seed transmission in faba bean, pea, and chickpea. Eur. J. Plant Pathol. 1992;98:329–342. doi: 10.1007/BF01974461. - DOI
-
- Fortass M., Diallo S. Broad bean mottle virus in Morocco; curculionid vectors, and natural occurrence in food legumes other than faba bean (Vicia faba) Eur. J. Plant Pathol. 1993;99:219–226. doi: 10.1007/BF01974666. - DOI
-
- Makkouk K.M., Bos L., Rizkallah A., Azzam O.I., Katul L. Broad bean mottle virus: Identification, host range, serology, and occurrence on faba bean (Vicia faba) in West Asia and North Africa. Eur. J. Plant Pathol. 1989;94:195–212. doi: 10.1007/BF02006546. - DOI
-
- Bawden F.C., Chaudhuri R.P., Kassanis B. Some properties of broad-bean mottle virus. Ann. Appl. Biol. 1951;38:774–784. doi: 10.1111/j.1744-7348.1951.tb07849.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

