Cancer-Associated Dysregulation of Sumo Regulators: Proteases and Ligases
- PMID: 35887358
- PMCID: PMC9316396
- DOI: 10.3390/ijms23148012
Cancer-Associated Dysregulation of Sumo Regulators: Proteases and Ligases
Abstract
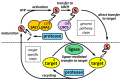
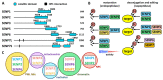
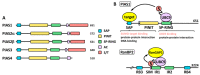
SUMOylation is a post-translational modification that has emerged in recent decades as a mechanism involved in controlling diverse physiological processes and that is essential in vertebrates. The SUMO pathway is regulated by several enzymes, proteases and ligases being the main actors involved in the control of sumoylation of specific targets. Dysregulation of the expression, localization and function of these enzymes produces physiological changes that can lead to the appearance of different types of cancer, depending on the enzymes and target proteins involved. Among the most studied proteases and ligases, those of the SENP and PIAS families stand out, respectively. While the proteases involved in this pathway have specific SUMO activity, the ligases may have additional functions unrelated to sumoylation, which makes it more difficult to study their SUMO-associated role in cancer process. In this review we update the knowledge and advances in relation to the impact of dysregulation of SUMO proteases and ligases in cancer initiation and progression.
Keywords: SUMO; cancer; ligase; protease; regulation; transcription.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
The Role of SUMO E3 Ligases in Signaling Pathway of Cancer Cells.Int J Mol Sci. 2022 Mar 26;23(7):3639. doi: 10.3390/ijms23073639. Int J Mol Sci. 2022. PMID: 35408996 Free PMC article. Review.
-
Mapping the SUMOylated landscape.FEBS J. 2015 Oct;282(19):3669-80. doi: 10.1111/febs.13378. Epub 2015 Jul 31. FEBS J. 2015. PMID: 26185901 Free PMC article. Review.
-
SUMOylation of ATF3 alters its transcriptional activity on regulation of TP53 gene.J Cell Biochem. 2013 Mar;114(3):589-98. doi: 10.1002/jcb.24396. J Cell Biochem. 2013. PMID: 22991139
-
Function and regulation of SUMO proteases.Nat Rev Mol Cell Biol. 2012 Dec;13(12):755-66. doi: 10.1038/nrm3478. Nat Rev Mol Cell Biol. 2012. PMID: 23175280 Free PMC article. Review.
-
Molecular mechanisms in SUMO conjugation.Biochem Soc Trans. 2020 Feb 28;48(1):123-135. doi: 10.1042/BST20190357. Biochem Soc Trans. 2020. PMID: 31872228 Review.
Cited by
-
Impacts of Nutlin-3a and exercise on murine double minute 2-enriched glioma treatment.Neural Regen Res. 2025 Apr 1;20(4):1135-1152. doi: 10.4103/NRR.NRR-D-23-00875. Epub 2024 Mar 1. Neural Regen Res. 2025. PMID: 38989952 Free PMC article.
-
SENP6-Mediated deSUMOylation of VEGFR2 Enhances Its Cell Membrane Transport in Angiogenesis.Int J Mol Sci. 2023 Jan 29;24(3):2544. doi: 10.3390/ijms24032544. Int J Mol Sci. 2023. PMID: 36768878 Free PMC article.
-
Exploring the Role of Unconventional Post-Translational Modifications in Cancer Diagnostics and Therapy.Curr Protein Pept Sci. 2024;25(10):780-796. doi: 10.2174/0113892037274615240528113148. Curr Protein Pept Sci. 2024. PMID: 38910429 Review.
-
Discovery of a Dual SENP1 and SENP2 Inhibitor.Int J Mol Sci. 2022 Oct 11;23(20):12085. doi: 10.3390/ijms232012085. Int J Mol Sci. 2022. PMID: 36292935 Free PMC article.
-
Understanding the Combined Effects of High Glucose Induced Hyper-Osmotic Stress and Oxygen Tension in the Progression of Tumourigenesis: From Mechanism to Anti-Cancer Therapeutics.Cells. 2023 Mar 7;12(6):825. doi: 10.3390/cells12060825. Cells. 2023. PMID: 36980166 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

