Construction of a lncRNA-mRNA Co-Expression Network for Nasopharyngeal Carcinoma
- PMID: 35875165
- PMCID: PMC9302896
- DOI: 10.3389/fonc.2022.809760
Construction of a lncRNA-mRNA Co-Expression Network for Nasopharyngeal Carcinoma
Abstract
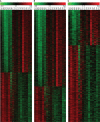
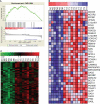
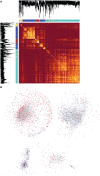
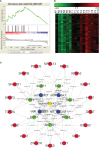
Long non-coding RNAs (lncRNAs) widely regulate gene expression and play important roles in the pathogenesis of human diseases, including malignant tumors. However, the functions of most lncRNAs remain to be elucidated. In order to study and screen novel lncRNAs with important functions in the carcinogenesis of nasopharyngeal carcinoma (NPC), we constructed a lncRNA expression profile of 10 NPC tissues and 6 controls through a gene microarray. We identified 1,276 lncRNAs, of which most are unknown, with different expression levels in the healthy and NPC tissues. In order to shed light on the functions of these unknown lncRNAs, we first constructed a co-expression network of lncRNAs and mRNAs using bioinformatics and systematic biological approach. Moreover, mRNAs were clustered and enriched by their biological functions, and those lncRNAs have similar expression trends with mRNAs were defined as functional molecules with potential biological significance. The module may help identify key lncRNAs in the carcinogenesis of NPC and provide clues for in-depth study of their functions and associated signaling pathways. We suggest the newly identified lncRNAs may have clinic value as biomarkers and therapeutic targets for NPC diagnosis and treatment.
Keywords: MYC; genomic instability; long non-coding RNA; nasopharyngeal carcinoma; p53; weighted gene co-expression network analysis.
Copyright © 2022 Fan, Xiong, Tang, Li, Zhu, Mo, Wang, Zhang, Gong, Liao, Li, Zeng, Guo, Xiong and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma.Tumour Biol. 2016 Mar;37(3):3683-95. doi: 10.1007/s13277-015-4156-x. Epub 2015 Oct 13. Tumour Biol. 2016. PMID: 26462838
-
Genome-wide analysis of long non-coding RNA in primary nasopharyngeal carcinoma by microarray.Histopathology. 2015 Jun;66(7):1022-30. doi: 10.1111/his.12616. Epub 2015 Jan 15. Histopathology. 2015. PMID: 25406670
-
Microarray Expression Profiling of Long Non-Coding RNAs Involved in Nasopharyngeal Carcinoma Metastasis.Int J Mol Sci. 2016 Nov 23;17(11):1956. doi: 10.3390/ijms17111956. Int J Mol Sci. 2016. PMID: 27886062 Free PMC article.
-
Functions and Roles of Long-Non-Coding RNAs in Human Nasopharyngeal Carcinoma.Cell Physiol Biochem. 2018;45(3):1191-1204. doi: 10.1159/000487451. Epub 2018 Feb 9. Cell Physiol Biochem. 2018. PMID: 29448252 Review.
-
Emerging roles of lncRNA in Nasopharyngeal Carcinoma and therapeutic opportunities.Int J Biol Sci. 2022 Mar 28;18(7):2714-2728. doi: 10.7150/ijbs.70292. eCollection 2022. Int J Biol Sci. 2022. PMID: 35541920 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

