Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1
- PMID: 35871297
- PMCID: PMC9371903
- DOI: 10.1093/nar/gkac620
Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1
Abstract
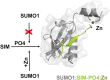
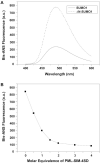
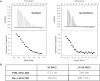
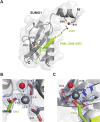
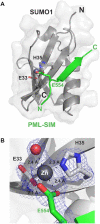
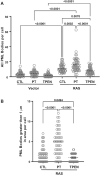
SUMO proteins are important regulators of many key cellular functions in part through their ability to form interactions with other proteins containing SUMO interacting motifs (SIMs). One characteristic feature of all SUMO proteins is the presence of a highly divergent intrinsically disordered region at their N-terminus. In this study, we examine the role of this N-terminal region of SUMO proteins in SUMO-SIM interactions required for the formation of nuclear bodies by the promyelocytic leukemia (PML) protein (PML-NBs). We demonstrate that the N-terminal region of SUMO1 functions in a paralog specific manner as an auto-inhibition domain by blocking its binding to the phosphorylated SIMs of PML and Daxx. Interestingly, we find that this auto-inhibition in SUMO1 is relieved by zinc, and structurally show that zinc stabilizes the complex between SUMO1 and a phospho-mimetic form of the SIM of PML. In addition, we demonstrate that increasing cellular zinc levels enhances PML-NB formation in senescent cells. Taken together, these results provide important insights into a paralog specific function of SUMO1, and suggest that zinc levels could play a crucial role in regulating SUMO1-SIM interactions required for PML-NB formation and function.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner.Structure. 2020 Feb 4;28(2):157-168.e5. doi: 10.1016/j.str.2019.11.019. Epub 2019 Dec 23. Structure. 2020. PMID: 31879127
-
Structural and functional characterization of the phosphorylation-dependent interaction between PML and SUMO1.Structure. 2015 Jan 6;23(1):126-138. doi: 10.1016/j.str.2014.10.015. Epub 2014 Dec 11. Structure. 2015. PMID: 25497731
-
SUMO Ligase Protein Inhibitor of Activated STAT1 (PIAS1) Is a Constituent Promyelocytic Leukemia Nuclear Body Protein That Contributes to the Intrinsic Antiviral Immune Response to Herpes Simplex Virus 1.J Virol. 2016 Jun 10;90(13):5939-5952. doi: 10.1128/JVI.00426-16. Print 2016 Jul 1. J Virol. 2016. PMID: 27099310 Free PMC article.
-
A Tale of Usurpation and Subversion: SUMO-Dependent Integrity of Promyelocytic Leukemia Nuclear Bodies at the Crossroad of Infection and Immunity.Front Cell Dev Biol. 2021 Aug 27;9:696234. doi: 10.3389/fcell.2021.696234. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34513832 Free PMC article. Review.
-
PML Body Biogenesis: A Delicate Balance of Interactions.Int J Mol Sci. 2023 Nov 24;24(23):16702. doi: 10.3390/ijms242316702. Int J Mol Sci. 2023. PMID: 38069029 Free PMC article. Review.
Cited by
-
Regulating the p53 Tumor Suppressor Network at PML Biomolecular Condensates.Cancers (Basel). 2022 Sep 20;14(19):4549. doi: 10.3390/cancers14194549. Cancers (Basel). 2022. PMID: 36230470 Free PMC article. Review.
-
SUMOylation at the crossroads of gut health: insights into physiology and pathology.Cell Commun Signal. 2024 Aug 19;22(1):404. doi: 10.1186/s12964-024-01786-5. Cell Commun Signal. 2024. PMID: 39160548 Free PMC article. Review.
References
-
- Desterro J.M., Rodriguez M.S., Kemp G.D., Hay R.T.. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 1999; 274:10618–10624. - PubMed
-
- Pichler A., Gast A., Seeler J.S., Dejean A., Melchior F.. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002; 108:109–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

