ADAR2 enzymes: efficient site-specific RNA editors with gene therapy aspirations
- PMID: 35863867
- PMCID: PMC9479739
- DOI: 10.1261/rna.079266.122
ADAR2 enzymes: efficient site-specific RNA editors with gene therapy aspirations
Abstract
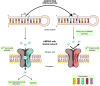
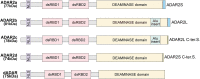
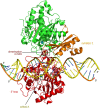
The adenosine deaminase acting on RNA (ADAR) enzymes are essential for neuronal function and innate immune control. ADAR1 RNA editing prevents aberrant activation of antiviral dsRNA sensors through editing of long, double-stranded RNAs (dsRNAs). In this review, we focus on the ADAR2 proteins involved in the efficient, highly site-specific RNA editing to recode open reading frames first discovered in the GRIA2 transcript encoding the key GLUA2 subunit of AMPA receptors; ADAR1 proteins also edit many of these sites. We summarize the history of ADAR2 protein research and give an up-to-date review of ADAR2 structural studies, human ADARBI (ADAR2) mutants causing severe infant seizures, and mouse disease models. Structural studies on ADARs and their RNA substrates facilitate current efforts to develop ADAR RNA editing gene therapy to edit disease-causing single nucleotide polymorphisms (SNPs). Artificial ADAR guide RNAs are being developed to retarget ADAR RNA editing to new target transcripts in order to correct SNP mutations in them at the RNA level. Site-specific RNA editing has been expanded to recode hundreds of sites in CNS transcripts in Drosophila and cephalopods. In Drosophila and C. elegans, ADAR RNA editing also suppresses responses to self dsRNA.
Keywords: ADAR; ADARB1; dsRNA; neurons; recoding RNA editing.
© 2022 Hajji et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures
Similar articles
-
Adenosine Deaminase Acting on RNA (ADAR) Enzymes: A Journey from Weird to Wondrous.Acc Chem Res. 2023 Nov 21;56(22):3165-3174. doi: 10.1021/acs.accounts.3c00433. Epub 2023 Oct 31. Acc Chem Res. 2023. PMID: 37906879 Free PMC article.
-
ADAR RNA editing in innate immune response phasing, in circadian clocks and in sleep.Biochim Biophys Acta Gene Regul Mech. 2019 Mar;1862(3):356-369. doi: 10.1016/j.bbagrm.2018.10.011. Epub 2018 Oct 31. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30391332 Review.
-
A comparative analysis of ADAR mutant mice reveals site-specific regulation of RNA editing.RNA. 2020 Apr;26(4):454-469. doi: 10.1261/rna.072728.119. Epub 2020 Jan 15. RNA. 2020. PMID: 31941663 Free PMC article.
-
Functional conservation in human and Drosophila of Metazoan ADAR2 involved in RNA editing: loss of ADAR1 in insects.Nucleic Acids Res. 2011 Sep 1;39(16):7249-62. doi: 10.1093/nar/gkr423. Epub 2011 May 27. Nucleic Acids Res. 2011. PMID: 21622951 Free PMC article.
-
ADAR RNA editing in human disease; more to it than meets the I.Hum Genet. 2017 Sep;136(9):1265-1278. doi: 10.1007/s00439-017-1837-0. Epub 2017 Sep 14. Hum Genet. 2017. PMID: 28913566 Review.
Cited by
-
RNA Editing-Dependent and -Independent Roles of Adenosine Deaminases Acting on RNA Proteins in Herpesvirus Infection-Hints on Another Layer of Complexity.Viruses. 2023 Sep 27;15(10):2007. doi: 10.3390/v15102007. Viruses. 2023. PMID: 37896783 Free PMC article. Review.
-
Small nucleolar RNA host gene 18 controls vascular smooth muscle cell contractile phenotype and neointimal hyperplasia.Cardiovasc Res. 2024 May 29;120(7):796-810. doi: 10.1093/cvr/cvae055. Cardiovasc Res. 2024. PMID: 38498586 Free PMC article.
-
A pipeline for identifying guide RNA sequences that promote RNA editing of nonsense mutations that cause inherited retinal diseases.Mol Ther Nucleic Acids. 2024 Jan 30;35(1):102130. doi: 10.1016/j.omtn.2024.102130. eCollection 2024 Mar 12. Mol Ther Nucleic Acids. 2024. PMID: 38375504 Free PMC article.
-
RNA editing enzymes: structure, biological functions and applications.Cell Biosci. 2024 Mar 16;14(1):34. doi: 10.1186/s13578-024-01216-6. Cell Biosci. 2024. PMID: 38493171 Free PMC article. Review.
-
Revealing the hidden RBP-RNA interactions with RNA modification enzyme-based strategies.Wiley Interdiscip Rev RNA. 2024 May-Jun;15(3):e1863. doi: 10.1002/wrna.1863. Wiley Interdiscip Rev RNA. 2024. PMID: 39392204 Free PMC article. Review.
References
-
- Ali IM, Evehe MSB, Netongo PM, Atogho-Tiedeu B, Akindeh-Nji M, Ngora H, Domkam IK, Diakite M, Baldip K, Ranford-Cartwright L, et al. 2014. Host candidate gene polymorphisms and associated clearance of P. falciparum amodiaquine and fansidar resistance mutants in children less than 5 years in Cameroon. Pathog Glob Health 108: 323–333. 10.1179/2047773214Y.0000000159 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
