Kinesin-8-specific loop-2 controls the dual activities of the motor domain according to tubulin protofilament shape
- PMID: 35859148
- PMCID: PMC9300613
- DOI: 10.1038/s41467-022-31794-3
Kinesin-8-specific loop-2 controls the dual activities of the motor domain according to tubulin protofilament shape
Abstract
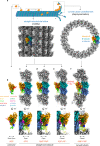
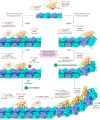
Kinesin-8s are dual-activity motor proteins that can move processively on microtubules and depolymerize microtubule plus-ends, but their mechanism of combining these distinct activities remains unclear. We addressed this by obtaining cryo-EM structures (2.6-3.9 Å) of Candida albicans Kip3 in different catalytic states on the microtubule lattice and on a curved microtubule end mimic. We also determined a crystal structure of microtubule-unbound CaKip3-ADP (2.0 Å) and analyzed the biochemical activity of CaKip3 and kinesin-1 mutants. These data reveal that the microtubule depolymerization activity of kinesin-8 originates from conformational changes of its motor core that are amplified by dynamic contacts between its extended loop-2 and tubulin. On curved microtubule ends, loop-1 inserts into preceding motor domains, forming head-to-tail arrays of kinesin-8s that complement loop-2 contacts with curved tubulin and assist depolymerization. On straight tubulin protofilaments in the microtubule lattice, loop-2-tubulin contacts inhibit conformational changes in the motor core, but in the ADP-Pi state these contacts are relaxed, allowing neck-linker docking for motility. We propose that these tubulin shape-induced alternations between pro-microtubule-depolymerization and pro-motility kinesin states, regulated by loop-2, are the key to the dual activity of kinesin-8 motors.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
New insights into the mechanochemical coupling mechanism of kinesin-microtubule complexes from their high-resolution structures.Biochem Soc Trans. 2023 Aug 31;51(4):1505-1520. doi: 10.1042/BST20221238. Biochem Soc Trans. 2023. PMID: 37560910 Free PMC article. Review.
-
Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s.Nat Commun. 2018 Apr 25;9(1):1662. doi: 10.1038/s41467-018-04044-8. Nat Commun. 2018. PMID: 29695795 Free PMC article.
-
A Tubulin Binding Switch Underlies Kip3/Kinesin-8 Depolymerase Activity.Dev Cell. 2017 Jul 10;42(1):37-51.e8. doi: 10.1016/j.devcel.2017.06.011. Dev Cell. 2017. PMID: 28697331 Free PMC article.
-
Structural model of microtubule dynamics inhibition by kinesin-4 from the crystal structure of KLP-12 -tubulin complex.Elife. 2022 Sep 6;11:e77877. doi: 10.7554/eLife.77877. Elife. 2022. PMID: 36065637 Free PMC article.
-
Kinesin, 30 years later: Recent insights from structural studies.Protein Sci. 2015 Jul;24(7):1047-56. doi: 10.1002/pro.2697. Epub 2015 Jun 11. Protein Sci. 2015. PMID: 25975756 Free PMC article. Review.
Cited by
-
New insights into the mechanochemical coupling mechanism of kinesin-microtubule complexes from their high-resolution structures.Biochem Soc Trans. 2023 Aug 31;51(4):1505-1520. doi: 10.1042/BST20221238. Biochem Soc Trans. 2023. PMID: 37560910 Free PMC article. Review.
-
Two Tetrahymena kinesin-9 family members exhibit slow plus-end-directed motility in vitro.Sci Rep. 2024 Sep 9;14(1):20993. doi: 10.1038/s41598-024-71280-y. Sci Rep. 2024. PMID: 39251704 Free PMC article.
-
Nucleotide-free structures of KIF20A illuminate atypical mechanochemistry in this kinesin-6.Open Biol. 2023 Sep;13(9):230122. doi: 10.1098/rsob.230122. Epub 2023 Sep 20. Open Biol. 2023. PMID: 37726093 Free PMC article.
-
Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L.Nat Commun. 2024 Jul 2;15(1):5530. doi: 10.1038/s41467-024-48720-4. Nat Commun. 2024. PMID: 38956021 Free PMC article.
-
Magic-angle-spinning NMR structure of the kinesin-1 motor domain assembled with microtubules reveals the elusive neck linker orientation.Nat Commun. 2022 Nov 10;13(1):6795. doi: 10.1038/s41467-022-34026-w. Nat Commun. 2022. PMID: 36357375 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

