Uncovering the enigmatic evolution of bears in greater depth: The hybrid origin of the Asiatic black bear
- PMID: 35858381
- PMCID: PMC9351369
- DOI: 10.1073/pnas.2120307119
Uncovering the enigmatic evolution of bears in greater depth: The hybrid origin of the Asiatic black bear
Abstract
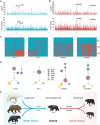
Bears are fascinating mammals because of their complex pattern of speciation and rapid evolution of distinct phenotypes. Interspecific hybridization has been common and has shaped the complex evolutionary history of bears. In this study, based on the largest population-level genomic dataset to date involving all Ursinae species and recently developed methods for detecting hybrid speciation, we provide explicit evidence for the hybrid origin of Asiatic black bears, which arose through historical hybridization between the ancestor of polar bear/brown bear/American black bears and the ancestor of sun bear/sloth bears. This was inferred to have occurred soon after the divergence of the two parental lineages in Eurasia due to climate-driven population expansion and dispersal. In addition, we found that the intermediate body size of this hybrid species arose from its combination of relevant genes derived from two parental lineages of contrasting sizes. This and alternate fixation of numerous other loci that had diverged between parental lineages may have initiated the reproductive isolation of the Asiatic black bear from its two parents. Our study sheds further light on the evolutionary history of bears and documents the importance of hybridization in new species formation and phenotypic evolution in mammals.
Keywords: Asiatic black bear; Ursinae; hybridization; population genomics.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
The evolutionary history of bears is characterized by gene flow across species.Sci Rep. 2017 Apr 19;7:46487. doi: 10.1038/srep46487. Sci Rep. 2017. PMID: 28422140 Free PMC article.
-
High genetic diversity and distinct ancient lineage of Asiatic black bears revealed by non-invasive surveys in the Annapurna Conservation Area, Nepal.PLoS One. 2018 Dec 5;13(12):e0207662. doi: 10.1371/journal.pone.0207662. eCollection 2018. PLoS One. 2018. PMID: 30517155 Free PMC article.
-
Genomic evidence for island population conversion resolves conflicting theories of polar bear evolution.PLoS Genet. 2013;9(3):e1003345. doi: 10.1371/journal.pgen.1003345. Epub 2013 Mar 14. PLoS Genet. 2013. PMID: 23516372 Free PMC article.
-
Evaluating hybrid speciation and swamping in wild carnivores with a decision-tree approach.Conserv Biol. 2024 Feb;38(1):e14197. doi: 10.1111/cobi.14197. Epub 2023 Nov 22. Conserv Biol. 2024. PMID: 37811741 Review.
-
EPIDEMIOLOGIC AND PUBLIC HEALTH SIGNIFICANCE OF TOXOPLASMA GONDII INFECTIONS IN BEARS (URSUS SPP.): A 50 YEAR REVIEW INCLUDING RECENT GENETIC EVIDENCE.J Parasitol. 2021 May 1;107(3):519-528. doi: 10.1645/21-16. J Parasitol. 2021. PMID: 34167147 Review.
Cited by
-
Genomic signatures suggesting adaptation to ocean acidification in a coral holobiont from volcanic CO2 seeps.Commun Biol. 2023 Jul 22;6(1):769. doi: 10.1038/s42003-023-05103-7. Commun Biol. 2023. PMID: 37481685 Free PMC article.
-
Late Pleistocene polar bear genomes reveal the timing of allele fixation in key genes associated with Arctic adaptation.BMC Genomics. 2024 Sep 16;25(1):826. doi: 10.1186/s12864-024-10617-3. BMC Genomics. 2024. PMID: 39278943 Free PMC article.
-
Ancient DNA Reveals Maternal Philopatry of the Northeast Eurasian Brown Bear (Ursus arctos) Population during the Holocene.Genes (Basel). 2022 Oct 27;13(11):1961. doi: 10.3390/genes13111961. Genes (Basel). 2022. PMID: 36360198 Free PMC article.
-
A most aggressive bear: Safari videos document sloth bear defense against tiger predation.Ecol Evol. 2024 Jul 11;14(7):e11524. doi: 10.1002/ece3.11524. eCollection 2024 Jul. Ecol Evol. 2024. PMID: 39005887 Free PMC article.
-
Deep Ancestral Introgressions between Ovine Species Shape Sheep Genomes via Argali-Mediated Gene Flow.Mol Biol Evol. 2024 Nov 1;41(11):msae212. doi: 10.1093/molbev/msae212. Mol Biol Evol. 2024. PMID: 39404100 Free PMC article.
References
-
- Preuß A., Gansloßer U., Purschke G., Magiera U., Bear-hybrids: Behaviour and phenotype. Zool Garten N F 78, 204–220 (2009).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

