Epigenetic mechanisms and potential therapeutic targets in stroke
- PMID: 35854641
- PMCID: PMC9580166
- DOI: 10.1177/0271678X221116192
Epigenetic mechanisms and potential therapeutic targets in stroke
Abstract
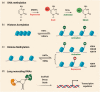
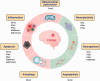
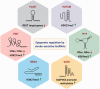
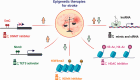
Accumulating evidence indicates a central role for epigenetic modifications in the progression of stroke pathology. These epigenetic mechanisms are involved in complex and dynamic processes that modulate post-stroke gene expression, cellular injury response, motor function, and cognitive ability. Despite decades of research, stroke continues to be classified as a leading cause of death and disability worldwide with limited clinical interventions. Thus, technological advances in the field of epigenetics may provide innovative targets to develop new stroke therapies. This review presents the evidence on the impact of epigenomic readers, writers, and erasers in both ischemic and hemorrhagic stroke pathophysiology. We specifically explore the role of DNA methylation, DNA hydroxymethylation, histone modifications, and epigenomic regulation by long non-coding RNAs in modulating gene expression and functional outcome after stroke. Furthermore, we highlight promising pharmacological approaches and biomarkers in relation to epigenetics for translational therapeutic applications.
Keywords: Biomarkers; cerebral ischemia; epigenomics; hemorrhagic stroke; neuroprotection.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures
Similar articles
-
Minor Changes for a Major Impact: A Review of Epigenetic Modifications in Cell-Based Therapies for Stroke.Int J Mol Sci. 2022 Oct 28;23(21):13106. doi: 10.3390/ijms232113106. Int J Mol Sci. 2022. PMID: 36361891 Free PMC article. Review.
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
Emerging role of epigenetics in stroke: part 1: DNA methylation and chromatin modifications.Arch Neurol. 2010 Nov;67(11):1316-22. doi: 10.1001/archneurol.2010.275. Arch Neurol. 2010. PMID: 21060009 Free PMC article. Review.
-
The Emerging Role of Epigenetics in Cerebral Ischemia.Mol Neurobiol. 2017 Apr;54(3):1887-1905. doi: 10.1007/s12035-016-9788-3. Epub 2016 Feb 19. Mol Neurobiol. 2017. PMID: 26894397 Review.
-
[Advances in epigenetics in ischemic stroke].Zhongguo Zhong Yao Za Zhi. 2022 Sep;47(17):4551-4559. doi: 10.19540/j.cnki.cjcmm.20220425.602. Zhongguo Zhong Yao Za Zhi. 2022. PMID: 36164859 Chinese.
Cited by
-
Targeting epigenetic and posttranslational modifications regulating ferroptosis for the treatment of diseases.Signal Transduct Target Ther. 2023 Dec 10;8(1):449. doi: 10.1038/s41392-023-01720-0. Signal Transduct Target Ther. 2023. PMID: 38072908 Free PMC article. Review.
-
Post-stroke depression: epigenetic and epitranscriptomic modifications and their interplay with gut microbiota.Mol Psychiatry. 2023 Oct;28(10):4044-4055. doi: 10.1038/s41380-023-02099-8. Epub 2023 May 15. Mol Psychiatry. 2023. PMID: 37188778 Free PMC article. Review.
-
Genomics of Brain Disorders 4.0.Int J Mol Sci. 2024 Mar 25;25(7):3667. doi: 10.3390/ijms25073667. Int J Mol Sci. 2024. PMID: 38612479 Free PMC article.
-
Inhibition of the JAK2/STAT3 pathway and cell cycle re-entry contribute to the protective effect of remote ischemic pre-conditioning of rat hindlimbs on cerebral ischemia/reperfusion injury.CNS Neurosci Ther. 2023 Mar;29(3):866-877. doi: 10.1111/cns.14023. Epub 2022 Nov 23. CNS Neurosci Ther. 2023. PMID: 36419252 Free PMC article.
-
Epigenetics as a target to mitigate excess stroke risk in people of African ancestry: A scoping review.J Stroke Cerebrovasc Dis. 2024 May;33(5):107585. doi: 10.1016/j.jstrokecerebrovasdis.2024.107585. Epub 2024 Jan 20. J Stroke Cerebrovasc Dis. 2024. PMID: 38253246
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

