The mechanism of replication stalling and recovery within repetitive DNA
- PMID: 35853874
- PMCID: PMC9296464
- DOI: 10.1038/s41467-022-31657-x
The mechanism of replication stalling and recovery within repetitive DNA
Abstract
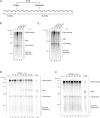
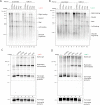
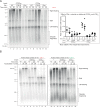
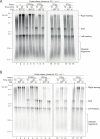
Accurate chromosomal DNA replication is essential to maintain genomic stability. Genetic evidence suggests that certain repetitive sequences impair replication, yet the underlying mechanism is poorly defined. Replication could be directly inhibited by the DNA template or indirectly, for example by DNA-bound proteins. Here, we reconstitute replication of mono-, di- and trinucleotide repeats in vitro using eukaryotic replisomes assembled from purified proteins. We find that structure-prone repeats are sufficient to impair replication. Whilst template unwinding is unaffected, leading strand synthesis is inhibited, leading to fork uncoupling. Synthesis through hairpin-forming repeats is rescued by replisome-intrinsic mechanisms, whereas synthesis of quadruplex-forming repeats requires an extrinsic accessory helicase. DNA-induced fork stalling is mechanistically similar to that induced by leading strand DNA lesions, highlighting structure-prone repeats as an important potential source of replication stress. Thus, we propose that our understanding of the cellular response to replication stress may also be applied to DNA-induced replication stalling.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Restarted replication forks are error-prone and cause CAG repeat expansions and contractions.PLoS Genet. 2021 Oct 21;17(10):e1009863. doi: 10.1371/journal.pgen.1009863. eCollection 2021 Oct. PLoS Genet. 2021. PMID: 34673780 Free PMC article.
-
Differential requirement of Srs2 helicase and Rad51 displacement activities in replication of hairpin-forming CAG/CTG repeats.Nucleic Acids Res. 2017 May 5;45(8):4519-4531. doi: 10.1093/nar/gkx088. Nucleic Acids Res. 2017. PMID: 28175398 Free PMC article.
-
Dynamics of Replication Fork Progression Following Helicase-Polymerase Uncoupling in Eukaryotes.J Mol Biol. 2019 May 3;431(10):2040-2049. doi: 10.1016/j.jmb.2019.03.011. Epub 2019 Mar 17. J Mol Biol. 2019. PMID: 30894292 Free PMC article.
-
Replisome structure suggests mechanism for continuous fork progression and post-replication repair.DNA Repair (Amst). 2019 Sep;81:102658. doi: 10.1016/j.dnarep.2019.102658. Epub 2019 Jul 8. DNA Repair (Amst). 2019. PMID: 31303546 Free PMC article. Review.
-
Contacts and context that regulate DNA helicase unwinding and replisome progression.Enzymes. 2019;45:183-223. doi: 10.1016/bs.enz.2019.08.001. Epub 2019 Sep 12. Enzymes. 2019. PMID: 31627877 Review.
Cited by
-
How Pol α-primase is targeted to replisomes to prime eukaryotic DNA replication.Mol Cell. 2023 Aug 17;83(16):2911-2924.e16. doi: 10.1016/j.molcel.2023.06.035. Epub 2023 Jul 27. Mol Cell. 2023. PMID: 37506699 Free PMC article.
-
Mechanisms for Maintaining Eukaryotic Replisome Progression in the Presence of DNA Damage.Front Mol Biosci. 2021 Jul 6;8:712971. doi: 10.3389/fmolb.2021.712971. eCollection 2021. Front Mol Biosci. 2021. PMID: 34295925 Free PMC article. Review.
-
A new G-quadruplex-specific photosensitizer inducing genome instability in cancer cells by triggering oxidative DNA damage and impeding replication fork progression.Nucleic Acids Res. 2023 Jul 7;51(12):6264-6285. doi: 10.1093/nar/gkad365. Nucleic Acids Res. 2023. PMID: 37191066 Free PMC article.
-
The fragile X locus is prone to spontaneous DNA damage that is preferentially repaired by nonhomologous end-joining to preserve genome integrity.iScience. 2024 Jan 6;27(2):108814. doi: 10.1016/j.isci.2024.108814. eCollection 2024 Feb 16. iScience. 2024. PMID: 38303711 Free PMC article.
-
Replication dependent and independent mechanisms of GAA repeat instability.DNA Repair (Amst). 2022 Oct;118:103385. doi: 10.1016/j.dnarep.2022.103385. Epub 2022 Aug 3. DNA Repair (Amst). 2022. PMID: 35952488 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

