Genome-Wide Detection of Copy Number Variations and Evaluation of Candidate Copy Number Polymorphism Genes Associated With Complex Traits of Pigs
- PMID: 35847642
- PMCID: PMC9280686
- DOI: 10.3389/fvets.2022.909039
Genome-Wide Detection of Copy Number Variations and Evaluation of Candidate Copy Number Polymorphism Genes Associated With Complex Traits of Pigs
Abstract
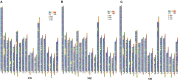
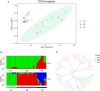
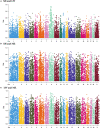
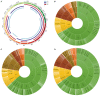
Copy number variation (CNV) has been considered to be an important source of genetic variation for important phenotypic traits of livestock. In this study, we performed whole-genome CNV detection on Suhuai (SH) (n = 23), Chinese Min Zhu (MZ) (n = 11), and Large White (LW) (n = 12) pigs based on next-generation sequencing data. The copy number variation regions (CNVRs) were annotated and analyzed, and 10,885, 10,836, and 10,917 CNVRs were detected in LW, MZ, and SH pigs, respectively. Some CNVRs have been randomly selected for verification of the variation type by real-time PCR. We found that SH and LW pigs are closely related, while MZ pigs are distantly related to the SH and LW pigs by CNVR-based genetic structure, PCA, VST, and QTL analyses. A total of 14 known genes annotated in CNVRs were unique for LW pigs. Among them, the cyclin T2 (CCNT2) is involved in cell proliferation and the cell cycle. The FA Complementation Group M (FANCM) is involved in defective DNA repair and reproductive cell development. Ten known genes annotated in 47 CNVRs were unique for MZ pigs. The genes included glycerol-3-phosphate acyltransferase 3 (GPAT3) is involved in fat synthesis and is essential to forming the glycerol triphosphate. Glutathione S-transferase mu 4 (GSTM4) gene plays an important role in detoxification. Eleven known genes annotated in 23 CNVRs were unique for SH pigs. Neuroligin 4 X-linked (NLGN4X) and Neuroligin 4 Y-linked (NLGN4Y) are involved with nerve disorders and nerve signal transmission. IgLON family member 5 (IGLON5) is related to autoimmunity and neural activities. The unique characteristics of LW, MZ, and SH pigs are related to these genes with CNV polymorphisms. These findings provide important information for the identification of candidate genes in the molecular breeding of pigs.
Keywords: crossbreeding; economic traits; evolution; genetic structure analysis; livestock.
Copyright © 2022 Zhang, Zhao, Guo, Xu, Liu, Cheng, Chao, Schinckel and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Genome-wide detection of CNV regions and their potential association with growth and fatness traits in Duroc pigs.BMC Genomics. 2021 May 8;22(1):332. doi: 10.1186/s12864-021-07654-7. BMC Genomics. 2021. PMID: 33964879 Free PMC article.
-
A comprehensive survey of copy number variation in 18 diverse pig populations and identification of candidate copy number variable genes associated with complex traits.BMC Genomics. 2012 Dec 27;13:733. doi: 10.1186/1471-2164-13-733. BMC Genomics. 2012. PMID: 23270433 Free PMC article.
-
Copy Number Variation and Selection Signal: Exploring the Domestication History and Phenotype Differences Between Duroc and the Chinese Native Ningxiang Pigs.Int J Mol Sci. 2024 Oct 31;25(21):11716. doi: 10.3390/ijms252111716. Int J Mol Sci. 2024. PMID: 39519268 Free PMC article.
-
Genome wide copy number variations using Porcine 60K SNP Beadchip in Landlly pigs.Anim Biotechnol. 2023 Nov;34(6):1891-1899. doi: 10.1080/10495398.2022.2056047. Epub 2022 Apr 4. Anim Biotechnol. 2023. PMID: 35369845 Review.
-
Copy number variation in livestock: A mini review.Vet World. 2018 Apr;11(4):535-541. doi: 10.14202/vetworld.2018.535-541. Epub 2018 Apr 26. Vet World. 2018. PMID: 29805222 Free PMC article. Review.
Cited by
-
Unveiling the Influence of Copy Number Variations on Genetic Diversity and Adaptive Evolution in China's Native Pig Breeds via Whole-Genome Resequencing.Int J Mol Sci. 2024 May 27;25(11):5843. doi: 10.3390/ijms25115843. Int J Mol Sci. 2024. PMID: 38892031 Free PMC article.
-
Genome-Wide Scan for Copy Number Variations in Chinese Merino Sheep Based on Ovine High-Density 600K SNP Arrays.Animals (Basel). 2024 Oct 8;14(19):2897. doi: 10.3390/ani14192897. Animals (Basel). 2024. PMID: 39409846 Free PMC article.
-
A composite strategy of genome-wide association study and copy number variation analysis for carcass traits in a Duroc pig population.BMC Genomics. 2022 Aug 13;23(1):590. doi: 10.1186/s12864-022-08804-1. BMC Genomics. 2022. PMID: 35964005 Free PMC article.
-
Copy Number Variation Analysis Revealed the Evolutionary Difference between Chinese Indigenous Pigs and Asian Wild Boars.Genes (Basel). 2023 Feb 12;14(2):472. doi: 10.3390/genes14020472. Genes (Basel). 2023. PMID: 36833399 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

