The Plant V-ATPase
- PMID: 35845650
- PMCID: PMC9280200
- DOI: 10.3389/fpls.2022.931777
The Plant V-ATPase
Abstract
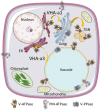
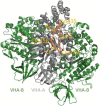
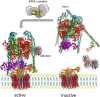
V-ATPase is the dominant proton pump in plant cells. It contributes to cytosolic pH homeostasis and energizes transport processes across endomembranes of the secretory pathway. Its localization in the trans Golgi network/early endosomes is essential for vesicle transport, for instance for the delivery of cell wall components. Furthermore, it is crucial for response to abiotic and biotic stresses. The V-ATPase's rather complex structure and multiple subunit isoforms enable high structural flexibility with respect to requirements for different organs, developmental stages, and organelles. This complexity further demands a sophisticated assembly machinery and transport routes in cells, a process that is still not fully understood. Regulation of V-ATPase is a target of phosphorylation and redox-modifications but also involves interactions with regulatory proteins like 14-3-3 proteins and the lipid environment. Regulation by reversible assembly, as reported for yeast and the mammalian enzyme, has not be proven in plants but seems to be absent in autotrophic cells. Addressing the regulation of V-ATPase is a promising approach to adjust its activity for improved stress resistance or higher crop yield.
Keywords: Arabidopsis; V-ATPase; glucose; pH-homeostasis; proton pump.
Copyright © 2022 Seidel.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
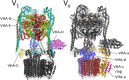

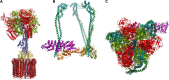
Similar articles
-
Regulation of V-ATPase Activity and Organelle pH by Phosphatidylinositol Phosphate Lipids.Front Cell Dev Biol. 2020 Jun 23;8:510. doi: 10.3389/fcell.2020.00510. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32656214 Free PMC article. Review.
-
The cellular biology of proton-motive force generation by V-ATPases.J Exp Biol. 2000 Jan;203(Pt 1):89-95. doi: 10.1242/jeb.203.1.89. J Exp Biol. 2000. PMID: 10600677 Review.
-
Vacuolar and plasma membrane proton-adenosinetriphosphatases.Physiol Rev. 1999 Apr;79(2):361-85. doi: 10.1152/physrev.1999.79.2.361. Physiol Rev. 1999. PMID: 10221984 Review.
-
Interaction between the yeast RAVE complex and Vph1-containing Vo sectors is a central glucose-sensitive interaction required for V-ATPase reassembly.J Biol Chem. 2020 Feb 21;295(8):2259-2269. doi: 10.1074/jbc.RA119.011522. Epub 2020 Jan 15. J Biol Chem. 2020. PMID: 31941791 Free PMC article.
-
Luminal and cytosolic pH feedback on proton pump activity and ATP affinity of V-type ATPase from Arabidopsis.J Biol Chem. 2012 Mar 16;287(12):8986-93. doi: 10.1074/jbc.M111.310367. Epub 2012 Jan 3. J Biol Chem. 2012. PMID: 22215665 Free PMC article.
Cited by
-
Regulation of V-ATPase by Jasmonic Acid: Possible Role of Persulfidation.Int J Mol Sci. 2023 Sep 9;24(18):13896. doi: 10.3390/ijms241813896. Int J Mol Sci. 2023. PMID: 37762199 Free PMC article.
-
Membrane Proteomics to Understand Enhancement Effects of Millimeter-Wave Irradiation on Wheat Root under Flooding Stress.Int J Mol Sci. 2023 May 19;24(10):9014. doi: 10.3390/ijms24109014. Int J Mol Sci. 2023. PMID: 37240359 Free PMC article.
-
Selection of Reference Genes and HSP17.9A Expression Profiling in Heat-Stressed Grapevine Varieties.Genes (Basel). 2024 Sep 30;15(10):1283. doi: 10.3390/genes15101283. Genes (Basel). 2024. PMID: 39457407 Free PMC article.
-
Human V-ATPase a-subunit isoforms bind specifically to distinct phosphoinositide phospholipids.J Biol Chem. 2023 Dec;299(12):105473. doi: 10.1016/j.jbc.2023.105473. Epub 2023 Nov 17. J Biol Chem. 2023. PMID: 37979916 Free PMC article.
-
Creating Climate-Resilient Crops by Increasing Drought, Heat, and Salt Tolerance.Plants (Basel). 2024 Apr 29;13(9):1238. doi: 10.3390/plants13091238. Plants (Basel). 2024. PMID: 38732452 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

