Estimating the timing of multiple admixture events using 3-locus linkage disequilibrium
- PMID: 35839249
- PMCID: PMC9342778
- DOI: 10.1371/journal.pgen.1010281
Estimating the timing of multiple admixture events using 3-locus linkage disequilibrium
Abstract
Estimating admixture histories is crucial for understanding the genetic diversity we see in present-day populations. Allele frequency or phylogeny-based methods are excellent for inferring the existence of admixture or its proportions. However, to estimate admixture times, spatial information from admixed chromosomes of local ancestry or the decay of admixture linkage disequilibrium (ALD) is used. One popular method, implemented in the programs ALDER and ROLLOFF, uses two-locus ALD to infer the time of a single admixture event, but is only able to estimate the time of the most recent admixture event based on this summary statistic. To address this limitation, we derive analytical expressions for the expected ALD in a three-locus system and provide a new statistical method based on these results that is able to resolve more complicated admixture histories. Using simulations, we evaluate the performance of this method on a range of different admixture histories. As an example, we apply the method to the Colombian and Mexican samples from the 1000 Genomes project. The implementation of our method is available at https://github.com/Genomics-HSE/LaNeta.
Conflict of interest statement
The authors have declared that no competing interests exist.
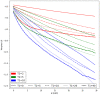
Figures

Similar articles
-
Inferring admixture histories of human populations using linkage disequilibrium.Genetics. 2013 Apr;193(4):1233-54. doi: 10.1534/genetics.112.147330. Epub 2013 Feb 14. Genetics. 2013. PMID: 23410830 Free PMC article.
-
Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium.Am J Hum Genet. 2001 Jan;68(1):198-207. doi: 10.1086/316935. Epub 2000 Dec 7. Am J Hum Genet. 2001. PMID: 11112661 Free PMC article.
-
Fast individual ancestry inference from DNA sequence data leveraging allele frequencies for multiple populations.BMC Bioinformatics. 2015 Jan 16;16:4. doi: 10.1186/s12859-014-0418-7. BMC Bioinformatics. 2015. PMID: 25592880 Free PMC article.
-
Admixture mapping and the role of population structure for localizing disease genes.Adv Genet. 2008;60:547-69. doi: 10.1016/S0065-2660(07)00419-1. Adv Genet. 2008. PMID: 18358332 Review.
-
Linkage disequilibrium mapping: the role of population history, size, and structure.Adv Genet. 2001;42:413-37. doi: 10.1016/s0065-2660(01)42034-7. Adv Genet. 2001. PMID: 11037333 Review.
Cited by
-
On the Distribution of Tract Lengths During Adaptive Introgression.G3 (Bethesda). 2020 Oct 5;10(10):3663-3673. doi: 10.1534/g3.120.401616. G3 (Bethesda). 2020. PMID: 32763953 Free PMC article.
-
Estimating the Timing of Multiple Admixture Pulses During Local Ancestry Inference.Genetics. 2018 Nov;210(3):1089-1107. doi: 10.1534/genetics.118.301411. Epub 2018 Sep 11. Genetics. 2018. PMID: 30206187 Free PMC article.
-
Unappreciated subcontinental admixture in Europeans and European Americans and implications for genetic epidemiology studies.Nat Commun. 2023 Nov 7;14(1):6802. doi: 10.1038/s41467-023-42491-0. Nat Commun. 2023. PMID: 37935687 Free PMC article.

