Metagenomic Next-Generation Sequencing Successfully Detects Pulmonary Infectious Pathogens in Children With Hematologic Malignancy
- PMID: 35837477
- PMCID: PMC9273861
- DOI: 10.3389/fcimb.2022.899028
Metagenomic Next-Generation Sequencing Successfully Detects Pulmonary Infectious Pathogens in Children With Hematologic Malignancy
Abstract
Background: Pulmonary infection is a leading cause of mortality in pediatric patients with hematologic malignancy (HM). In clinical settings, pulmonary pathogens are frequently undetectable, and empiric therapies may be costly, ineffective and lead to poor outcomes in this vulnerable population. Metagenomic next-generation sequencing (mNGS) enhances pathogen detection, but data on its application in pediatric patients with HM and pulmonary infections are scarce.
Methods: We retrospectively reviewed 55 pediatric patients with HM and pulmonary infection who were performed mNGS on bronchoalveolar lavage fluid from January 2020 to October 2021. The performances of mNGS methods and conventional microbiological methods in pathogenic diagnosis and subsequently antibiotic adjustment were investigated.
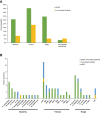
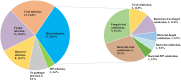
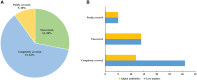
Results: A definite or probable microbial etiology of pulmonary infection was established for 50 of the 55 patients (90.9%) when mNGS was combined with conventional microbiological tests. The positive rate was 87.3% (48 of 55 patients) for mNGS versus 34.5% (19 of 55 patients) with conventional microbiological methods (P < 0.001). Bacteria, viruses and fungi were detected in 17/55 (30.9%), 25/55 (45.5%) and 19/55 (34.5%) cases using mNGS, respectively. Furthermore, 17 patients (30.9%) were identified as pulmonary mixed infections. Among the 50 pathogen-positive cases, 38% (19/50) were not completely pathogen-covered by empirical antibiotics and all of them were accordingly made an antibiotic adjustment. In the present study, the 30-day mortality rate was 7.3%.
Conclusion: mNGS is a valuable diagnostic tool to determine the etiology and appropriate treatment in pediatric patients with HM and pulmonary infection. In these vulnerable children with HM, pulmonary infections are life-threatening, so we recommend that mNGS should be considered as a front-line diagnostic test.
Keywords: children; diagnosis; hematologic malignancy; mNGS; pulmonary infection.
Copyright © 2022 Wang, Wang, Ding, Tang, Zhang, Chen and You.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Improving Suspected Pulmonary Infection Diagnosis by Bronchoalveolar Lavage Fluid Metagenomic Next-Generation Sequencing: a Multicenter Retrospective Study.Microbiol Spectr. 2022 Aug 31;10(4):e0247321. doi: 10.1128/spectrum.02473-21. Epub 2022 Aug 9. Microbiol Spectr. 2022. PMID: 35943274 Free PMC article.
-
Application of Metagenomic Next-Generation Sequencing (mNGS) Using Bronchoalveolar Lavage Fluid (BALF) in Diagnosing Pneumonia of Children.Microbiol Spectr. 2022 Oct 26;10(5):e0148822. doi: 10.1128/spectrum.01488-22. Epub 2022 Sep 28. Microbiol Spectr. 2022. PMID: 36169415 Free PMC article.
-
Diagnostic value of bronchoalveolar lavage fluid metagenomic next-generation sequencing in pediatric pneumonia.Front Cell Infect Microbiol. 2022 Oct 27;12:950531. doi: 10.3389/fcimb.2022.950531. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36389175 Free PMC article.
-
The clinical application of metagenomic next-generation sequencing for detecting pathogens in bronchoalveolar lavage fluid: case reports and literature review.Expert Rev Mol Diagn. 2022 May;22(5):575-582. doi: 10.1080/14737159.2022.2071607. Epub 2022 May 2. Expert Rev Mol Diagn. 2022. PMID: 35473493 Review.
-
Metagenomic next generation sequencing of bronchoalveolar lavage fluids for the identification of pathogens in patients with pulmonary infection: A retrospective study.Diagn Microbiol Infect Dis. 2024 Sep;110(1):116402. doi: 10.1016/j.diagmicrobio.2024.116402. Epub 2024 Jun 12. Diagn Microbiol Infect Dis. 2024. PMID: 38878340 Review.
Cited by
-
Diagnostic value of a nanopore sequencing assay of bronchoalveolar lavage fluid in pulmonary tuberculosis.BMC Pulm Med. 2023 Mar 8;23(1):77. doi: 10.1186/s12890-023-02337-3. BMC Pulm Med. 2023. PMID: 36890507 Free PMC article.
-
Nanopore sequencing for smear-negative pulmonary tuberculosis-a multicentre prospective study in China.Ann Clin Microbiol Antimicrob. 2024 Jun 14;23(1):51. doi: 10.1186/s12941-024-00714-2. Ann Clin Microbiol Antimicrob. 2024. PMID: 38877520 Free PMC article.
-
Clinical and diagnostic values of metagenomic next-generation sequencing for infection in hematology patients: a systematic review and meta-analysis.BMC Infect Dis. 2024 Feb 7;24(1):167. doi: 10.1186/s12879-024-09073-x. BMC Infect Dis. 2024. PMID: 38326763 Free PMC article.
-
[Value of metagenomic next-generation sequencing in children with hematological malignancies complicated with infections].Zhongguo Dang Dai Er Ke Za Zhi. 2023 Jul 15;25(7):718-725. doi: 10.7499/j.issn.1008-8830.2212059. Zhongguo Dang Dai Er Ke Za Zhi. 2023. PMID: 37529954 Free PMC article. Chinese.
References
-
- Chellapandian D., Lehrnbecher T., Phillips B., Fisher B., Zaoutis T., Steinbach W., et al. . (2015). Bronchoalveolar Lavage and Lung Biopsy in Patients With Cancer and Hematopoietic Stem-Cell Transplantation Recipients: A Systematic Review and Meta-Analysis. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 33, 501–509. doi: 10.1200/jco.2014.58.0480 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

