MicroRNA turnover: a tale of tailing, trimming, and targets
- PMID: 35811249
- PMCID: PMC9789169
- DOI: 10.1016/j.tibs.2022.06.005
MicroRNA turnover: a tale of tailing, trimming, and targets
Abstract
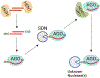
MicroRNAs (miRNAs) post-transcriptionally repress gene expression by guiding Argonaute (AGO) proteins to target mRNAs. While much is known about the regulation of miRNA biogenesis, miRNA degradation pathways are comparatively poorly understood. Although miRNAs generally exhibit slow turnover, they can be rapidly degraded through regulated mechanisms that act in a context- or sequence-specific manner. Recent work has revealed a particularly important role for specialized target interactions in controlling rates of miRNA degradation. Engagement of these targets is associated with the addition and removal of nucleotides from the 3' ends of miRNAs, a process known as tailing and trimming. Here we review these mechanisms of miRNA modification and turnover, highlighting the contexts in which they impact miRNA stability and discussing important questions that remain unanswered.
Keywords: Argonaute; TDMD; ZSWIM8; degradation; miRNA; tailing and trimming; target-directed miRNA degradation.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests J.T.M is a scientific advisor for Ribometrix, Inc. and Circ Bio, Inc.
Figures

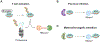

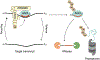
Similar articles
-
Target-directed microRNA degradation: Mechanisms, significance, and functional implications.Wiley Interdiscip Rev RNA. 2024 Mar-Apr;15(2):e1832. doi: 10.1002/wrna.1832. Wiley Interdiscip Rev RNA. 2024. PMID: 38448799 Free PMC article. Review.
-
A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming.Science. 2020 Dec 18;370(6523):eabc9546. doi: 10.1126/science.abc9546. Epub 2020 Nov 12. Science. 2020. PMID: 33184234 Free PMC article.
-
Structural Basis for Target-Directed MicroRNA Degradation.Mol Cell. 2019 Sep 19;75(6):1243-1255.e7. doi: 10.1016/j.molcel.2019.06.019. Epub 2019 Jul 25. Mol Cell. 2019. PMID: 31353209 Free PMC article.
-
Widespread microRNA degradation elements in target mRNAs can assist the encoded proteins.Genes Dev. 2021 Dec 1;35(23-24):1595-1609. doi: 10.1101/gad.348874.121. Epub 2021 Nov 24. Genes Dev. 2021. PMID: 34819352 Free PMC article.
-
To kill a microRNA: emerging concepts in target-directed microRNA degradation.Nucleic Acids Res. 2024 Feb 28;52(4):1558-1574. doi: 10.1093/nar/gkae003. Nucleic Acids Res. 2024. PMID: 38224449 Free PMC article. Review.
Cited by
-
Recent progress in miRNA biogenesis and decay.RNA Biol. 2024 Jan;21(1):1-8. doi: 10.1080/15476286.2023.2288741. Epub 2023 Nov 29. RNA Biol. 2024. PMID: 38031325 Free PMC article. Review.
-
Target-directed microRNA degradation: Mechanisms, significance, and functional implications.Wiley Interdiscip Rev RNA. 2024 Mar-Apr;15(2):e1832. doi: 10.1002/wrna.1832. Wiley Interdiscip Rev RNA. 2024. PMID: 38448799 Free PMC article. Review.
-
The guide RNA sequence dictates the slicing kinetics and conformational dynamics of the Argonaute silencing complex.bioRxiv [Preprint]. 2024 Jun 20:2023.10.15.562437. doi: 10.1101/2023.10.15.562437. bioRxiv. 2024. Update in: Mol Cell. 2024 Aug 8;84(15):2918-2934.e11. doi: 10.1016/j.molcel.2024.06.026 PMID: 38766062 Free PMC article. Updated. Preprint.
-
A Review of IsomiRs in Colorectal Cancer.Noncoding RNA. 2023 Jun 7;9(3):34. doi: 10.3390/ncrna9030034. Noncoding RNA. 2023. PMID: 37368334 Free PMC article. Review.
-
Perspective on adolescent psychiatric illness and emerging role of microRNAs as biomarkers of risk.J Psychiatry Neurosci. 2024 Aug 29;49(4):E282-E288. doi: 10.1503/jpn.240072. Print 2024 Jul-Aug. J Psychiatry Neurosci. 2024. PMID: 39209460 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

