Critical Roles of Circular RNA in Tumor Metastasis via Acting as a Sponge of miRNA/isomiR
- PMID: 35806027
- PMCID: PMC9267010
- DOI: 10.3390/ijms23137024
Critical Roles of Circular RNA in Tumor Metastasis via Acting as a Sponge of miRNA/isomiR
Abstract
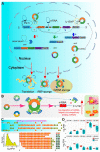
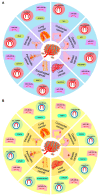
Circular RNAs (circRNAs), a class of new endogenous non-coding RNAs (ncRNAs), are closely related to the carcinogenic process and play a critical role in tumor metastasis. CircRNAs can lay the foundation for tumor metastasis via promoting tumor angiogenesis, make tumor cells gain the ability of migration and invasion by regulating epithelial-mesenchymal transition (EMT), interact with immune cells, cytokines, chemokines, and other non-cellular components in the tumor microenvironment, damage the normal immune function or escape the immunosuppressive network, and further promote cell survival and metastasis. Herein, based on the characteristics and biological functions of circRNA, we elaborated on the effect of circRNA via circRNA-associated competing endogenous RNA (ceRNA) network by acting as miRNA/isomiR sponges on tumor angiogenesis, cancer cell migration and invasion, and interaction with the tumor microenvironment (TME), then explored the potential interactions across different RNAs, and finally discussed the potential clinical value and application as a promising biomarker. These results provide a theoretical basis for the further application of metastasis-related circRNAs in cancer treatment. In summary, we briefly summarize the diverse roles of a circRNA-associated ceRNA network in cancer metastasis and the potential clinical application, especially the interaction of circRNA and miRNA/isomiR, which may complicate the RNA regulatory network and which will contribute to a novel insight into circRNA in the future.
Keywords: cancer metastasis; circular RNA (circRNA); competing endogenous RNA (ceRNA) network; isomiR; microRNA (miRNA).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
CircCCNB1 silencing acting as a miR-106b-5p sponge inhibited GPM6A expression to promote HCC progression by enhancing DYNC1I1 expression and activating the AKT/ERK signaling pathway.Int J Biol Sci. 2022 Jan 1;18(2):637-651. doi: 10.7150/ijbs.66915. eCollection 2022. Int J Biol Sci. 2022. PMID: 35002514 Free PMC article.
-
Identification of epithelial-mesenchymal transition-related circRNA-miRNA-mRNA ceRNA regulatory network in breast cancer.Pathol Res Pract. 2020 Sep;216(9):153088. doi: 10.1016/j.prp.2020.153088. Epub 2020 Jun 29. Pathol Res Pract. 2020. PMID: 32825956
-
Crosstalk in competing endogenous RNA networks reveals new circular RNAs involved in the pathogenesis of early HIV infection.J Transl Med. 2018 Nov 29;16(1):332. doi: 10.1186/s12967-018-1706-1. J Transl Med. 2018. PMID: 30486834 Free PMC article.
-
Biological functions and molecular mechanisms of exosome-derived circular RNAs and their clinical implications in digestive malignancies: the vintage in the bottle.Ann Med. 2024 Dec;56(1):2420861. doi: 10.1080/07853890.2024.2420861. Epub 2024 Nov 1. Ann Med. 2024. PMID: 39484707 Free PMC article. Review.
-
Epigenetic Associations between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma.Cancers (Basel). 2020 Sep 14;12(9):2622. doi: 10.3390/cancers12092622. Cancers (Basel). 2020. PMID: 32937886 Free PMC article. Review.
Cited by
-
The Tumorigenic Role of Circular RNA-MicroRNA Axis in Cancer.Int J Mol Sci. 2023 Feb 3;24(3):3050. doi: 10.3390/ijms24033050. Int J Mol Sci. 2023. PMID: 36769372 Free PMC article. Review.
-
Mechanism underlying circRNA dysregulation in the TME of digestive system cancer.Front Immunol. 2022 Sep 27;13:951561. doi: 10.3389/fimmu.2022.951561. eCollection 2022. Front Immunol. 2022. PMID: 36238299 Free PMC article. Review.
-
Therapeutic and diagnostic applications of exosomes in colorectal cancer.Med Oncol. 2024 Jul 20;41(8):203. doi: 10.1007/s12032-024-02440-3. Med Oncol. 2024. PMID: 39031221 Review.
-
CircGSK3β mediates PD-L1 transcription through miR-338-3p/PRMT5/H3K4me3 to promote breast cancer cell immune evasion and tumor progression.Cell Death Discov. 2024 Oct 4;10(1):426. doi: 10.1038/s41420-024-02197-8. Cell Death Discov. 2024. PMID: 39366935 Free PMC article.
-
Overexpression of circular RNA hsa_circ_0008621 facilitates colorectal cancer progression and predicts poor prognosis.Ann Gastroenterol Surg. 2024 Mar 22;8(4):639-649. doi: 10.1002/ags3.12793. eCollection 2024 Jul. Ann Gastroenterol Surg. 2024. PMID: 38957564 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

