Diversity of NKG2C genotypes in a European population: Conserved and recombinant haplotypes in the coding, promoter, and 3'-untranslated regions
- PMID: 35802353
- PMCID: PMC9796621
- DOI: 10.1111/tan.14734
Diversity of NKG2C genotypes in a European population: Conserved and recombinant haplotypes in the coding, promoter, and 3'-untranslated regions
Abstract
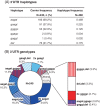
NK cells monitor altered molecular patterns in tumors and infected cells through an ample array of receptors. Two families of evolutionarily distant receptors have converged to enable human NK cells to sense levels of HLA class I ligands, frequently abnormal in altered cells. Whilst different forms of polymorphism are a hallmark of killer-cell immunoglobulin-like receptors and their classic HLA-A, B, and C ligands, genetic diversity of killer-cell lectin-like receptors for the non-classical HLA-E (CD94/NKG2 heterodimers) is less conspicuous and has attracted less attention. A common pattern of diversification in both receptor families is evolution of pairs of inhibitory and activating homologs for a common ligand, the genes encoding activating receptors being more frequently affected by copy number variation (CNV). This is exemplified by the gene encoding the activating NKG2C subunit (KLRC2 or NKG2C), which marks an NK-cell subpopulation that differentiates or expands in response to cytomegalovirus. We have studied NKG2C diversity in 240 South European individuals, using polymerase chain reaction and sequencing methods to assess both gene CNV and single-nucleotide polymorphisms (SNPs) affecting its promoter, coding and 3'-untranslated (3'UT) regions. Sequence analysis revealed eight common SNPs-one in the promoter, two in the coding sequence, and five in the 3'UT region. These SNPs associate strongly with each other, forming three conserved extended haplotypes (frequencies: 0.456, 0.221, and 0.117). Homo- and heterozygous combination of these, together with complete gene deletion (0.175) and additional haplotypes with frequencies lower than 0.015, generate a diversity of NKG2C genotypes of potential immunological importance.
Keywords: HLA-E; NKG2C receptor; alleles; genetic polymorphism; human genetics; natural killer cell lectin-like receptors.
© 2022 The Authors. HLA: Immune Response Genetics published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Deletion of the activating NKG2C receptor and a functional polymorphism in its ligand HLA-E in psoriasis susceptibility.Exp Dermatol. 2013 Oct;22(10):679-81. doi: 10.1111/exd.12233. Exp Dermatol. 2013. PMID: 24079744 Free PMC article.
-
The inhibitory NK cell receptor CD94/NKG2A and the activating receptor CD94/NKG2C bind the top of HLA-E through mostly shared but partly distinct sets of HLA-E residues.Eur J Immunol. 2004 Jan;34(1):81-90. doi: 10.1002/eji.200324432. Eur J Immunol. 2004. PMID: 14971033
-
Direct binding of purified HLA class I antigens by soluble NKG2/CD94 C-type lectins from natural killer cells.Scand J Immunol. 1999 May;49(5):459-65. doi: 10.1046/j.1365-3083.1999.00566.x. Scand J Immunol. 1999. PMID: 10320637
-
Diversification of both KIR and NKG2 natural killer cell receptor genes in macaques - implications for highly complex MHC-dependent regulation of natural killer cells.Immunology. 2017 Feb;150(2):139-145. doi: 10.1111/imm.12666. Epub 2016 Oct 5. Immunology. 2017. PMID: 27565739 Free PMC article. Review.
-
Activating NKG2C Receptor: Functional Characteristics and Current Strategies in Clinical Applications.Arch Immunol Ther Exp (Warsz). 2023 Mar 10;71(1):9. doi: 10.1007/s00005-023-00674-z. Arch Immunol Ther Exp (Warsz). 2023. PMID: 36899273 Free PMC article. Review.
Cited by
-
Polygenic polymorphism is associated with NKG2A repertoire and influences lymphocyte phenotype and function.Blood Adv. 2024 Oct 22;8(20):5382-5399. doi: 10.1182/bloodadvances.2024013508. Blood Adv. 2024. PMID: 39158076 Free PMC article.
-
Significance of HLA-E and its two NKG2 receptors in development of complications after allogeneic transplantation of hematopoietic stem cells.Front Immunol. 2023 Oct 13;14:1227897. doi: 10.3389/fimmu.2023.1227897. eCollection 2023. Front Immunol. 2023. PMID: 37901227 Free PMC article.
References
-
- Anderson SK, Ortaldo JR, McVicar DW. The ever‐expanding Ly49 gene family: repertoire and signaling. Immunol Rev. 2001;181:79‐89. - PubMed
-
- Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217‐251. - PubMed
-
- Braud VM, Allan DS, O'Callaghan CA, et al. HLA‐E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391(6669):795‐799. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

