Multiple modes of antigen exposure induce clonotypically diverse epitope-specific CD8+ T cells across multiple tissues in nonhuman primates
- PMID: 35797339
- PMCID: PMC9262242
- DOI: 10.1371/journal.ppat.1010611
Multiple modes of antigen exposure induce clonotypically diverse epitope-specific CD8+ T cells across multiple tissues in nonhuman primates
Abstract
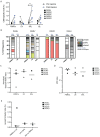
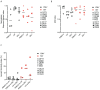
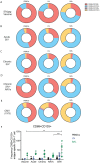
Antigen-specific CD8+ T cells play a key role in the host's antiviral response. T cells recognize viral epitopes via the T cell receptor (TCR), which contains the complementarity-determining region-3 (CDR3), comprising the variable, diversity and joining regions of the TCRβ gene. During chronic simian immunodeficiency virus (SIV) infection of Asian macaque nonhuman primates, tissue-specific clonotypes are identifiable among SIV-specific CD8+ T cells. Here, we sought to determine level of antigen exposure responsible for the tissue-specific clonotypic structure. We examined whether the priming event and/or chronic antigen exposure is response for tissue-specific TCR repertoires. We evaluated the TCR repertoire of SIV-specific CD8+ T cells after acute antigen exposure following inoculation with a SIV DNA vaccine, longitudinally during the acute and chronic phases of SIV, and after administration of antiretrovirals (ARVs). Finally, we assessed the TCR repertoire of cytomegalovirus (CMV)-specific CD8+ T cells to establish if TCR tissue-specificity is shared among viruses that chronically replicate. TCR sequences unique to anatomical sites were identified after acute antigen exposure via vaccination and upon acute SIV infection. Tissue-specific clones also persisted into chronic infection and the clonotypic structure continued to evolve after ARV administration. Finally, tissue-specific clones were also observed in CMV-specific CD8+ T cells. Together, these data suggest that acute antigen priming is sufficient to induce tissue-specific clones and that this clonal hierarchy can persist when antigen loads are naturally or therapeutically reduced, providing mechanistic insight into tissue-residency.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Multi-omics analysis of SIV-specific CD8+ T cells in multiple anatomical sites.PLoS Pathog. 2024 Sep 9;20(9):e1012545. doi: 10.1371/journal.ppat.1012545. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39250524 Free PMC article.
-
T Cell Receptor Diversity and Lineage Relationship between Virus-Specific CD8 T Cell Subsets during Chronic Lymphocytic Choriomeningitis Virus Infection.J Virol. 2020 Sep 29;94(20):e00935-20. doi: 10.1128/JVI.00935-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32759317 Free PMC article.
-
HIV-1 Coreceptor CXCR4 Antagonists Promote Clonal Expansion of Viral Epitope-Specific CD8+ T Cells During Acute SIV Infection in Rhesus Monkeys In Vivo.J Acquir Immune Defic Syndr. 2015 Jun 1;69(2):145-53. doi: 10.1097/QAI.0000000000000586. J Acquir Immune Defic Syndr. 2015. PMID: 25714247
-
Abundant cytomegalovirus (CMV) reactive clonotypes in the CD8(+) T cell receptor alpha repertoire following allogeneic transplantation.Clin Exp Immunol. 2016 Jun;184(3):389-402. doi: 10.1111/cei.12770. Epub 2016 Mar 8. Clin Exp Immunol. 2016. PMID: 26800118 Free PMC article.
-
Clonal repertoires of virus-specific CD8+ T lymphocytes are shared in mucosal and systemic compartments during chronic simian immunodeficiency virus infection in rhesus monkeys.J Immunol. 2010 Aug 15;185(4):2191-9. doi: 10.4049/jimmunol.1001340. Epub 2010 Jul 12. J Immunol. 2010. PMID: 20624939
Cited by
-
TCR repertoire landscape reveals macrophage-mediated clone deletion in endotoxin tolerance.Inflamm Res. 2023 Mar;72(3):531-540. doi: 10.1007/s00011-022-01685-w. Epub 2023 Jan 12. Inflamm Res. 2023. PMID: 36633616 Free PMC article.
-
Immunotoxin-mediated depletion of Gag-specific CD8+ T cells undermines natural control of SIV.JCI Insight. 2024 Jun 17;9(14):e174168. doi: 10.1172/jci.insight.174168. JCI Insight. 2024. PMID: 38885329 Free PMC article.
-
Antigen-specificity measurements are the key to understanding T cell responses.Front Immunol. 2023 Apr 14;14:1127470. doi: 10.3389/fimmu.2023.1127470. eCollection 2023. Front Immunol. 2023. PMID: 37122719 Free PMC article. Review.
-
Multi-omics analysis of SIV-specific CD8+ T cells in multiple anatomical sites.PLoS Pathog. 2024 Sep 9;20(9):e1012545. doi: 10.1371/journal.ppat.1012545. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39250524 Free PMC article.
-
Improved DNA Vaccine Delivery with Needle-Free Injection Systems.Vaccines (Basel). 2023 Jan 28;11(2):280. doi: 10.3390/vaccines11020280. Vaccines (Basel). 2023. PMID: 36851159 Free PMC article. Review.
References
-
- Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, et al.. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog. 2010;6(1):e1000747. Epub 2010/02/04. doi: 10.1371/journal.ppat.1000747 . - DOI - PMC - PubMed
-
- Veazey RS, Gauduin MC, Mansfield KG, Tham IC, Altman JD, Lifson JD, et al.. Emergence and kinetics of simian immunodeficiency virus-specific CD8(+) T cells in the intestines of macaques during primary infection. J Virol. 2001;75(21):10515–9. Epub 2001/10/03. doi: 10.1128/JVI.75.21.10515-10519.2001 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

