Genome-Wide Comprehensive Survey of the Subtilisin-Like Proteases Gene Family Associated With Rice Caryopsis Development
- PMID: 35795345
- PMCID: PMC9251471
- DOI: 10.3389/fpls.2022.943184
Genome-Wide Comprehensive Survey of the Subtilisin-Like Proteases Gene Family Associated With Rice Caryopsis Development
Abstract
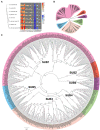
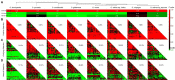
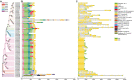
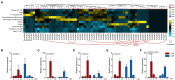
Subtilisin-like proteases (SUBs), which are extensively distributed in three life domains, affect all aspects of the plant life cycle, from embryogenesis and organogenesis to senescence. To explore the role of SUBs in rice caryopsis development, we recharacterized the OsSUB gene family in rice (Oryza sativa ssp. japonica). In addition, investigation of the SUBs was conducted across cultivated and wild rice in seven other Oryza diploid species (O. brachyantha, O. glaberrima, O. meridionalis, O. nivara, O. punctata, O. rufipogon, and O. sativa ssp. indica). Sixty-two OsSUBs were identified in the latest O. sativa ssp. japonica genome, which was higher than that observed in wild species. The SUB gene family was classified into six evolutionary branches, and SUB1 and SUB3 possessed all tandem duplication (TD) genes. All paralogous SUBs in eight Oryza plants underwent significant purifying selection. The expansion of SUBs in cultivated rice was primarily associated with the occurrence of tandem duplication events and purifying selection and may be the result of rice domestication. Combining the expression patterns of OsSUBs in different rice tissues and qRT-PCR verification, four OsSUBs were expressed in rice caryopses. Moreover, OsSUBs expressed in rice caryopses possessed an earlier origin in Oryza, and the gene cluster formed by OsSUBs together with the surrounding gene blocks may be responsible for the specific expression of OsSUBs in caryopses. All the above insights were inseparable from the continuous evolution and domestication of Oryza. Together, our findings not only contribute to the understanding of the evolution of SUBs in cultivated and wild rice but also lay the molecular foundation of caryopsis development and engineering improvement of crop yield.
Keywords: Oryza; expression in rice caryopsis; gene family; rice; subtilisin-like proteases.
Copyright © 2022 Zheng, Pang, Xue, Gao, Zhao, Wang and Han.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Genetic Conservation of CBS Domain Containing Protein Family in Oryza Species and Their Association with Abiotic Stress Responses.Int J Mol Sci. 2022 Feb 1;23(3):1687. doi: 10.3390/ijms23031687. Int J Mol Sci. 2022. PMID: 35163610 Free PMC article.
-
Genome-Wide Identification and Evolution Analysis of the CYP76 Subfamily in Rice (Oryza sativa).Int J Mol Sci. 2023 May 10;24(10):8522. doi: 10.3390/ijms24108522. Int J Mol Sci. 2023. PMID: 37239869 Free PMC article.
-
Evolutionary Analysis of GH3 Genes in Six Oryza Species/Subspecies and Their Expression under Salinity Stress in Oryza sativa ssp. japonica.Plants (Basel). 2019 Jan 24;8(2):30. doi: 10.3390/plants8020030. Plants (Basel). 2019. PMID: 30682815 Free PMC article.
-
New insights into the history of rice domestication.Trends Genet. 2007 Nov;23(11):578-87. doi: 10.1016/j.tig.2007.08.012. Epub 2007 Oct 25. Trends Genet. 2007. PMID: 17963977 Review.
-
The Genomics of Oryza Species Provides Insights into Rice Domestication and Heterosis.Annu Rev Plant Biol. 2019 Apr 29;70:639-665. doi: 10.1146/annurev-arplant-050718-100320. Annu Rev Plant Biol. 2019. PMID: 31035826 Review.
Cited by
-
Genome-Wide Identification of the AGC Protein Kinase Gene Family Related to Photosynthesis in Rice (Oryza sativa).Int J Mol Sci. 2022 Oct 19;23(20):12557. doi: 10.3390/ijms232012557. Int J Mol Sci. 2022. PMID: 36293407 Free PMC article.
-
Maize GOLDEN2-LIKE proteins enhance drought tolerance in rice by promoting stomatal closure.Plant Physiol. 2024 Jan 31;194(2):774-786. doi: 10.1093/plphys/kiad561. Plant Physiol. 2024. PMID: 37850886 Free PMC article.
References
LinkOut - more resources
Full Text Sources

