Notch signaling determines cell-fate specification of the two main types of vomeronasal neurons of rodents
- PMID: 35781337
- PMCID: PMC9340558
- DOI: 10.1242/dev.200448
Notch signaling determines cell-fate specification of the two main types of vomeronasal neurons of rodents
Abstract
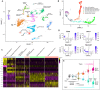
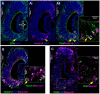
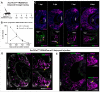
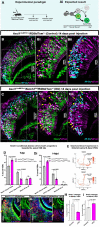
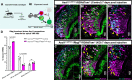
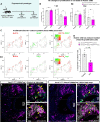
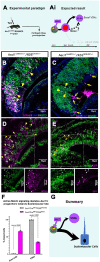
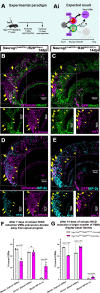
The ability of terrestrial vertebrates to find food and mating partners, and to avoid predators, relies on the detection of chemosensory information. Semiochemicals responsible for social and sexual behaviors are detected by chemosensory neurons of the vomeronasal organ (VNO), which transmits information to the accessory olfactory bulb. The vomeronasal sensory epithelium of most mammalian species contains a uniform vomeronasal system; however, rodents and marsupials have developed a more complex binary vomeronasal system, containing vomeronasal sensory neurons (VSNs) expressing receptors of either the V1R or V2R family. In rodents, V1R/apical and V2R/basal VSNs originate from a common pool of progenitors. Using single cell RNA-sequencing, we identified differential expression of Notch1 receptor and Dll4 ligand between the neuronal precursors at the VSN differentiation dichotomy. Our experiments show that Notch signaling is required for effective differentiation of V2R/basal VSNs. In fact, Notch1 loss of function in neuronal progenitors diverts them to the V1R/apical fate, whereas Notch1 gain of function redirects precursors to V2R/basal. Our results indicate that Notch signaling plays a pivotal role in triggering the binary differentiation dichotomy in the VNO of rodents.
Keywords: Dll4; Mouse; Neuronal dichotomy; Neuronal differentiation; Notch signaling; Single cell RNA sequencing; Vomeronasal organ.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

Similar articles
-
Bcl11b/Ctip2 controls the differentiation of vomeronasal sensory neurons in mice.J Neurosci. 2011 Jul 13;31(28):10159-73. doi: 10.1523/JNEUROSCI.1245-11.2011. J Neurosci. 2011. PMID: 21752992 Free PMC article.
-
The transcription factor Tfap2e/AP-2ε plays a pivotal role in maintaining the identity of basal vomeronasal sensory neurons.Dev Biol. 2018 Sep 1;441(1):67-82. doi: 10.1016/j.ydbio.2018.06.007. Epub 2018 Jun 19. Dev Biol. 2018. PMID: 29928868
-
Coordinated coexpression of two vomeronasal receptor V2R genes per neuron in the mouse.Mol Cell Neurosci. 2011 Feb;46(2):397-408. doi: 10.1016/j.mcn.2010.11.002. Epub 2010 Nov 26. Mol Cell Neurosci. 2011. PMID: 21112400
-
Mechanisms underlying pre- and postnatal development of the vomeronasal organ.Cell Mol Life Sci. 2021 Jun;78(12):5069-5082. doi: 10.1007/s00018-021-03829-3. Epub 2021 Apr 19. Cell Mol Life Sci. 2021. PMID: 33871676 Free PMC article. Review.
-
Signaling mechanisms and behavioral function of the mouse basal vomeronasal neuroepithelium.Front Neuroanat. 2014 Nov 26;8:135. doi: 10.3389/fnana.2014.00135. eCollection 2014. Front Neuroanat. 2014. PMID: 25505388 Free PMC article. Review.
Cited by
-
Phosphodiesterase 5A regulates the vomeronasal pump in mice.Genesis. 2024 Jun;62(3):e23603. doi: 10.1002/dvg.23603. Genesis. 2024. PMID: 38738564
-
Molecular, Cellular, and Developmental Organization of the Mouse Vomeronasal organ at Single Cell Resolution.bioRxiv [Preprint]. 2024 Sep 5:2024.02.22.581574. doi: 10.1101/2024.02.22.581574. bioRxiv. 2024. Update in: Elife. 2024 Dec 10;13:RP97356. doi: 10.7554/eLife.97356 PMID: 39253476 Free PMC article. Updated. Preprint.
-
Defective jagged-1 signaling affects GnRH development and contributes to congenital hypogonadotropic hypogonadism.JCI Insight. 2023 Mar 8;8(5):e161998. doi: 10.1172/jci.insight.161998. JCI Insight. 2023. PMID: 36729644 Free PMC article.
-
Illuminating the terminal nerve: Uncovering the link between GnRH-1 neuron and olfactory development.J Comp Neurol. 2024 Mar;532(3):e25599. doi: 10.1002/cne.25599. J Comp Neurol. 2024. PMID: 38488687
-
A revised conceptual framework for mouse vomeronasal pumping and stimulus sampling.Curr Biol. 2024 Mar 25;34(6):1206-1221.e6. doi: 10.1016/j.cub.2024.01.036. Epub 2024 Feb 5. Curr Biol. 2024. PMID: 38320553
References
-
- Aydin, B., Kakumanu, A., Rossillo, M., Moreno-Estellés, M., Garipler, G., Ringstad, N., Flames, N., Mahony, S. and Mazzoni, E. O. (2019). Proneural factors Ascl1 and Neurog2 contribute to neuronal subtype identities by establishing distinct chromatin landscapes. Nat. Neurosci. 22, 897-908. 10.1038/s41593-019-0399-y - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

