Comparison of temporal evolution of computed tomography imaging features in COVID-19 and influenza infections in a multicenter cohort study
- PMID: 35765661
- PMCID: PMC9226197
- DOI: 10.1016/j.ejro.2022.100431
Comparison of temporal evolution of computed tomography imaging features in COVID-19 and influenza infections in a multicenter cohort study
Abstract
Purpose: To compare temporal evolution of imaging features of coronavirus disease 2019 (COVID-19) and influenza in computed tomography and evaluate their predictive value for distinction.
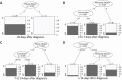
Methods: In this retrospective, multicenter study 179 CT examinations of 52 COVID-19 and 44 influenza critically ill patients were included. Lung involvement, main pattern (ground glass opacity, crazy paving, consolidation) and additional lung and chest findings were evaluated by two independent observers. Additional findings and clinical data were compared patient-wise. A decision tree analysis was performed to identify imaging features with predictive value in distinguishing both entities.
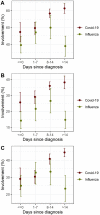
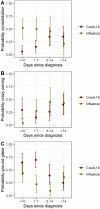
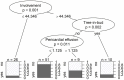
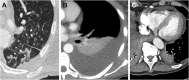
Results: In contrast to influenza patients, lung involvement remains high in COVID-19 patients > 14 days after the diagnosis. The predominant pattern in COVID-19 evolves from ground glass at the beginning to consolidation in later disease. In influenza there is more consolidation at the beginning and overall less ground glass opacity (p = 0.002). Decision tree analysis yielded the following: Earlier in disease course, pleural effusion is a typical feature of influenza (p = 0.007) whereas ground glass opacities indicate COVID-19 (p = 0.04). In later disease, particularly more lung involvement (p < 0.001), but also less pleural (p = 0.005) and pericardial (p = 0.003) effusion favor COVID-19 over influenza. Regardless of time point, less lung involvement (p < 0.001), tree-in-bud (p = 0.002) and pericardial effusion (p = 0.01) make influenza more likely than COVID-19.
Conclusions: This study identified differences in temporal evolution of imaging features between COVID-19 and influenza. These findings may help to distinguish both diseases in critically ill patients when laboratory findings are delayed or inconclusive.
Keywords: COPD, Chronic obstructive pulmonary disease; COVID-19; COVID-19, Coronavirus disease 2019; CT, Computed tomography; Computed tomography; GGO, Ground glass opacity; HIV, Human immunodeficiency virus; HSCT, Haematopoietic stem cell transplantation; ICC, Intraclass correlation coefficient; ICU, Intensive care unit; IQR, Interquartile range; Influenza; Lung; PCR, Polymerase chain reaction; Pneumonia; SD, Standard deviation; SOT, Solid organ transplantation.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures
Similar articles
-
[Spatial and temporal distribution and predictive value of chest CT scoring in patients with COVID-19].Zhonghua Jie He He Hu Xi Za Zhi. 2021 Mar 12;44(3):230-236. doi: 10.3760/cma.j.cn112147-20200522-00626. Zhonghua Jie He He Hu Xi Za Zhi. 2021. PMID: 33721937 Chinese.
-
COVID-19 Infection in Octagenarian Patients: Imaging Aspects and Clinical Correlations.Gerontology. 2022;68(3):261-271. doi: 10.1159/000516166. Epub 2021 Aug 5. Gerontology. 2022. PMID: 34515129 Free PMC article.
-
A Comparison of Clinical and Chest CT Findings in Patients With Influenza A (H1N1) Virus Infection and Coronavirus Disease (COVID-19).AJR Am J Roentgenol. 2020 Nov;215(5):1065-1071. doi: 10.2214/AJR.20.23214. Epub 2020 May 26. AJR Am J Roentgenol. 2020. PMID: 32452731
-
Role of Chest CT in COVID-19.J Clin Imaging Sci. 2021 Jun 3;11:30. doi: 10.25259/JCIS_138_2020. eCollection 2021. J Clin Imaging Sci. 2021. PMID: 34221639 Free PMC article. Review.
-
Computed tomography scan in COVID-19: a systematic review and meta-analysis.Pol J Radiol. 2022 Jan 5;87:e1-e23. doi: 10.5114/pjr.2022.112613. eCollection 2022. Pol J Radiol. 2022. PMID: 35140824 Free PMC article. Review.
Cited by
-
A Cluster of Paragonimiasis with Delayed Diagnosis Due to Difficulty Distinguishing Symptoms from Post-COVID-19 Respiratory Symptoms: A Report of Five Cases.Medicina (Kaunas). 2023 Jan 10;59(1):137. doi: 10.3390/medicina59010137. Medicina (Kaunas). 2023. PMID: 36676761 Free PMC article.
References
-
- Deng L.-S., Yuan J., Ding L., Chen Y.-L., Zhao C.-H., Chen G.-Q., Li X.-H., Li X.-H., Luo W.-T., Lan J.-F., Tan G.-Y., Tang S.-H., Xia J.-Y., Liu X. Comparison of patients hospitalized with COVID-19, H7N9 and H1N1. Infect. Dis. Poverty. 2020;9:163. doi: 10.1186/s40249-020-00781-5. - DOI - PMC - PubMed
-
- Zhou M., Yang D., Chen Y., Xu Y., Xu J.-F., Jie Z., Yao W., Jin X., Pan Z., Tan J., Wang L., Xia Y., Zou L., Xu X., Wei J., Guan M., Yan F., Feng J., Zhang H., Qu J. Deep learning for differentiating novel coronavirus pneumonia and influenza pneumonia. Ann. Transl. Med. 2021;9:111. doi: 10.21037/atm-20-5328. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

