Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics
- PMID: 35752612
- PMCID: PMC9233671
- DOI: 10.1038/s41392-022-01056-1
Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics
Abstract
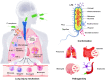
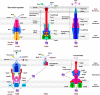
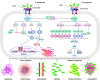
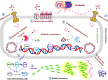
Pseudomonas aeruginosa (P. aeruginosa) is a Gram-negative opportunistic pathogen that infects patients with cystic fibrosis, burn wounds, immunodeficiency, chronic obstructive pulmonary disorder (COPD), cancer, and severe infection requiring ventilation, such as COVID-19. P. aeruginosa is also a widely-used model bacterium for all biological areas. In addition to continued, intense efforts in understanding bacterial pathogenesis of P. aeruginosa including virulence factors (LPS, quorum sensing, two-component systems, 6 type secretion systems, outer membrane vesicles (OMVs), CRISPR-Cas and their regulation), rapid progress has been made in further studying host-pathogen interaction, particularly host immune networks involving autophagy, inflammasome, non-coding RNAs, cGAS, etc. Furthermore, numerous technologic advances, such as bioinformatics, metabolomics, scRNA-seq, nanoparticles, drug screening, and phage therapy, have been used to improve our understanding of P. aeruginosa pathogenesis and host defense. Nevertheless, much remains to be uncovered about interactions between P. aeruginosa and host immune responses, including mechanisms of drug resistance by known or unannotated bacterial virulence factors as well as mammalian cell signaling pathways. The widespread use of antibiotics and the slow development of effective antimicrobials present daunting challenges and necessitate new theoretical and practical platforms to screen and develop mechanism-tested novel drugs to treat intractable infections, especially those caused by multi-drug resistance strains. Benefited from has advancing in research tools and technology, dissecting this pathogen's feature has entered into molecular and mechanistic details as well as dynamic and holistic views. Herein, we comprehensively review the progress and discuss the current status of P. aeruginosa biophysical traits, behaviors, virulence factors, invasive regulators, and host defense patterns against its infection, which point out new directions for future investigation and add to the design of novel and/or alternative therapeutics to combat this clinically significant pathogen.
© 2022. The Author(s).
Conflict of interest statement
The authors have no financial conflict of interest. Xiangrong Song and Min Wu are editorial board members of
Figures


Similar articles
-
The Small RNA ErsA Plays a Role in the Regulatory Network of Pseudomonas aeruginosa Pathogenicity in Airway Infections.mSphere. 2020 Oct 14;5(5):e00909-20. doi: 10.1128/mSphere.00909-20. mSphere. 2020. PMID: 33055260 Free PMC article.
-
TpiA is a Key Metabolic Enzyme That Affects Virulence and Resistance to Aminoglycoside Antibiotics through CrcZ in Pseudomonas aeruginosa.mBio. 2020 Jan 7;11(1):e02079-19. doi: 10.1128/mBio.02079-19. mBio. 2020. PMID: 31911486 Free PMC article.
-
Recent advances in therapeutic targets identification and development of treatment strategies towards Pseudomonas aeruginosa infections.BMC Microbiol. 2023 Mar 30;23(1):86. doi: 10.1186/s12866-023-02832-x. BMC Microbiol. 2023. PMID: 36991325 Free PMC article. Review.
-
Pseudomonas aeruginosa Leucine Aminopeptidase Influences Early Biofilm Composition and Structure via Vesicle-Associated Antibiofilm Activity.mBio. 2019 Nov 19;10(6):e02548-19. doi: 10.1128/mBio.02548-19. mBio. 2019. PMID: 31744920 Free PMC article.
-
Role of quorum sensing by Pseudomonas aeruginosa in microbial keratitis and cystic fibrosis.Microbiology (Reading). 2008 Aug;154(Pt 8):2184-2194. doi: 10.1099/mic.0.2008/019281-0. Microbiology (Reading). 2008. PMID: 18667552 Review.
Cited by
-
Acetylomics reveals an extensive acetylation diversity within Pseudomonas aeruginosa.Microlife. 2024 Sep 14;5:uqae018. doi: 10.1093/femsml/uqae018. eCollection 2024. Microlife. 2024. PMID: 39464744 Free PMC article.
-
In Vitro Digestion of Vacuum-Impregnated Yam Bean Snacks: Pediococcus acidilactici Viability and Mango Seed Polyphenol Bioaccessibility.Microorganisms. 2024 Sep 30;12(10):1993. doi: 10.3390/microorganisms12101993. Microorganisms. 2024. PMID: 39458302 Free PMC article.
-
Antibiofilm and Antimicrobial Potentials of Novel Synthesized Sulfur Camphor Derivatives.Int J Mol Sci. 2024 Oct 10;25(20):10895. doi: 10.3390/ijms252010895. Int J Mol Sci. 2024. PMID: 39456678 Free PMC article.
-
Impact of Nutrient Starvation on Biofilm Formation in Pseudomonas aeruginosa: An Analysis of Growth, Adhesion, and Spatial Distribution.Antibiotics (Basel). 2024 Oct 18;13(10):987. doi: 10.3390/antibiotics13100987. Antibiotics (Basel). 2024. PMID: 39452253 Free PMC article.
-
Molecular Epidemiology of Pseudomonas aeruginosa in Brazil: A Systematic Review and Meta-Analysis.Antibiotics (Basel). 2024 Oct 17;13(10):983. doi: 10.3390/antibiotics13100983. Antibiotics (Basel). 2024. PMID: 39452249 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

