The M13 Phage Assembly Machine Has a Membrane-Spanning Oligomeric Ring Structure
- PMID: 35746635
- PMCID: PMC9228878
- DOI: 10.3390/v14061163
The M13 Phage Assembly Machine Has a Membrane-Spanning Oligomeric Ring Structure
Abstract
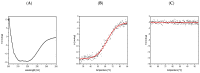
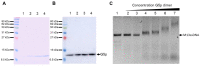
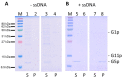
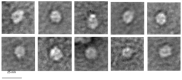
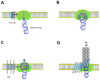
Bacteriophage M13 assembles its progeny particles in the inner membrane of the host. The major component of the assembly machine is G1p and together with G11p it generates an oligomeric structure with a pore-like inner cavity and an ATP hydrolysing domain. This allows the formation of the phage filament, which assembles multiple copies of the membrane-inserted major coat protein G8p around the extruding single-stranded circular DNA. The phage filament then passes through the G4p secretin that is localized in the outer membrane. Presumably, the inner membrane G1p/G11p and the outer G4p form a common complex. To unravel the structural details of the M13 assembly machine, we purified G1p from infected E. coli cells. The protein was overproduced together with G11p and solubilized from the membrane as a multimeric complex with a size of about 320 kDa. The complex revealed a pore-like structure with an outer diameter of about 12 nm, matching the dimensions of the outer membrane G4p secretin. The function of the M13 assembly machine for phage generation and secretion is discussed.
Keywords: affinity chromatography; bacteriophage M13; circular dichroism; membrane protein; phage assembly machine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Cysteine residues in the transmembrane regions of M13 procoat protein suggest that oligomeric coat proteins assemble onto phage progeny.J Bacteriol. 2007 Apr;189(7):2897-905. doi: 10.1128/JB.01551-06. Epub 2007 Jan 19. J Bacteriol. 2007. PMID: 17237167 Free PMC article.
-
Membrane insertion and assembly of epitope-tagged gp9 at the tip of the M13 phage.BMC Microbiol. 2011 Sep 26;11:211. doi: 10.1186/1471-2180-11-211. BMC Microbiol. 2011. PMID: 21943062 Free PMC article.
-
Design and evolution of artificial M13 coat proteins.J Mol Biol. 2000 Jun 30;300(1):213-9. doi: 10.1006/jmbi.2000.3845. J Mol Biol. 2000. PMID: 10864510
-
Engineering M13 for phage display.Biomol Eng. 2001 Sep;18(2):57-63. doi: 10.1016/s1389-0344(01)00087-9. Biomol Eng. 2001. PMID: 11535417 Review.
-
Protein-lipid interactions of bacteriophage M13 gene 9 minor coat protein.Mol Membr Biol. 2004 Nov-Dec;21(6):351-9. doi: 10.1080/09687860400012918. Mol Membr Biol. 2004. PMID: 15764365 Review.
Cited by
-
Cryo-electron microscopy of the f1 filamentous phage reveals insights into viral infection and assembly.Nat Commun. 2023 May 11;14(1):2724. doi: 10.1038/s41467-023-37915-w. Nat Commun. 2023. PMID: 37169795 Free PMC article.
-
Structural mechanisms of Tad pilus assembly and its interaction with an RNA virus.Sci Adv. 2024 May 3;10(18):eadl4450. doi: 10.1126/sciadv.adl4450. Epub 2024 May 3. Sci Adv. 2024. PMID: 38701202 Free PMC article.
References
-
- Kuhn A., Leptihn S. Encyclopedia of Virology. Elsevier; Amsterdam, The Netherlands: 2021. Helical and Filamentous Phages; pp. 53–60.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

