Homology Modelling, Molecular Docking and Molecular Dynamics Simulation Studies of CALMH1 against Secondary Metabolites of Bauhinia variegata to Treat Alzheimer's Disease
- PMID: 35741655
- PMCID: PMC9220886
- DOI: 10.3390/brainsci12060770
Homology Modelling, Molecular Docking and Molecular Dynamics Simulation Studies of CALMH1 against Secondary Metabolites of Bauhinia variegata to Treat Alzheimer's Disease
Abstract
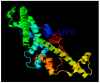
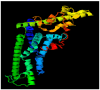
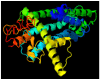
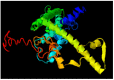
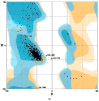
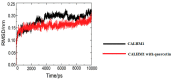
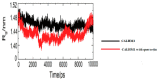
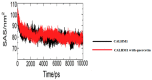
Calcium homeostasis modulator 1 (CALHM1) is a protein responsible for causing Alzheimer's disease. In the absence of an experimentally designed protein molecule, homology modelling was performed. Through homology modelling, different CALHM1 models were generated and validated through Rampage. To carry out further in silico studies, through molecular docking and molecular dynamics simulation experiments, various flavonoids and alkaloids from Bauhinia variegata were utilised as inhibitors to target the protein (CALHM1). The sequence of CALHM1 was retrieved from UniProt and the secondary structure prediction of CALHM1 was done through CFSSP, GOR4, and SOPMA methods. The structure was identified through LOMETS, MUSTER, and MODELLER and finally, the structures were validated through Rampage. Bauhinia variegata plant was used to check the interaction of alkaloids and flavonoids against CALHM1. The protein and protein-ligand complex were also validated through molecular dynamics simulations studies. The model generated through MODELLER software with 6VAM A was used because this model predicted the best results in the Ramachandran plot. Further molecular docking was performed, quercetin was found to be the most appropriate candidate for the protein molecule with the minimum binding energy of -12.45 kcal/mol and their ADME properties were analysed through Molsoft and Molinspiration. Molecular dynamics simulations showed that CALHM1 and CALHM1-quercetin complex became stable at 2500 ps. It may be seen through the study that quercetin may act as a good inhibitor for treatment. With the help of an in silico study, it was easier to analyse the 3D structure of the protein, which may be scrutinized for the best-predicted model. Quercetin may work as a good inhibitor for treating Alzheimer's disease, according to in silico research using molecular docking and molecular dynamics simulations, and future in vitro and in vivo analysis may confirm its effectiveness.
Keywords: AutoDock vina; LOMETS; MUSTER; homology modelling; iGEMDOCK.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






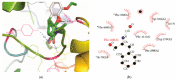
Similar articles
-
Screening and identification of secondary metabolites in the bark of Bauhinia variegata to treat Alzheimer's disease by using molecular docking and molecular dynamics simulations.J Biomol Struct Dyn. 2021 Oct;39(16):5988-5998. doi: 10.1080/07391102.2020.1796798. Epub 2020 Jul 28. J Biomol Struct Dyn. 2021. PMID: 32720564
-
Structure prediction of transferrin receptor protein 1 (TfR1) by homology modelling, docking, and molecular dynamics simulation studies.Heliyon. 2020 Jan 29;6(1):e03221. doi: 10.1016/j.heliyon.2020.e03221. eCollection 2020 Jan. Heliyon. 2020. PMID: 32021925 Free PMC article.
-
In silico QSAR analysis of quercetin reveals its potential as therapeutic drug for Alzheimer's disease.J Young Pharm. 2013 Dec;5(4):173-9. doi: 10.1016/j.jyp.2013.11.005. Epub 2013 Dec 15. J Young Pharm. 2013. PMID: 24563598 Free PMC article.
-
Synthesis, Molecular Docking, Molecular Dynamics Studies, and Biological Evaluation of 4H-Chromone-1,2,3,4-tetrahydropyrimidine-5-carboxylate Derivatives as Potential Antileukemic Agents.J Chem Inf Model. 2017 Jun 26;57(6):1246-1257. doi: 10.1021/acs.jcim.6b00138. Epub 2017 May 25. J Chem Inf Model. 2017. PMID: 28524659
-
Molecular Modeling Strategies of Cancer Multidrug Resistance.Drug Resist Updat. 2021 Dec;59:100789. doi: 10.1016/j.drup.2021.100789. Epub 2021 Nov 24. Drug Resist Updat. 2021. PMID: 34973929 Review.
Cited by
-
Biological Synthesis, Characterization, and Therapeutic Potential of S. commune-Mediated Gold Nanoparticles.Biomolecules. 2023 Dec 13;13(12):1785. doi: 10.3390/biom13121785. Biomolecules. 2023. PMID: 38136655 Free PMC article.
-
Network Pharmacology Reveals Key Targets and Pathways of Madhuca longifolia for Potential Alzheimer's Disease Treatment.Cell Biochem Biophys. 2024 Sep;82(3):2727-2746. doi: 10.1007/s12013-024-01389-4. Epub 2024 Jul 15. Cell Biochem Biophys. 2024. PMID: 39009828
-
New Insights into Molecular Mechanisms Underlying Neurodegenerative Disorders.Brain Sci. 2022 Sep 3;12(9):1190. doi: 10.3390/brainsci12091190. Brain Sci. 2022. PMID: 36138926 Free PMC article.
-
Quercetin Antagonizes the Sedative Effects of Linalool, Possibly through the GABAergic Interaction Pathway.Molecules. 2023 Jul 24;28(14):5616. doi: 10.3390/molecules28145616. Molecules. 2023. PMID: 37513487 Free PMC article.
-
Multitargeted Virtual Screening and Molecular Simulation of Natural Product-like Compounds against GSK3β, NMDA-Receptor, and BACE-1 for the Management of Alzheimer's Disease.Pharmaceuticals (Basel). 2023 Apr 20;16(4):622. doi: 10.3390/ph16040622. Pharmaceuticals (Basel). 2023. PMID: 37111379 Free PMC article.
References
-
- Chandra P.M., Venkateshwar J. Biological evaluation of Schiff bases of new isatin derivatives for anti Alzheimer’s activity. [(accessed on 31 December 2020)];Asian J. Pharm. Clin. Res. 2014 7:114–117. Available online: https://innovareacademics.in/journals/index.php/ajpcr/article/view/966.
-
- Khare N., Maheshwari S.K., Jha A.K. Screening and identification of secondary metabolites in the bark of Bauhinia variegata to treat Alzheimer’s disease by using molecular docking and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2020;39:5988–5998. doi: 10.1080/07391102.2020.1796798. - DOI - PubMed
-
- Ma Z., Siebert A.P., Cheung K.H., Lee R.J., Johnson B., Cohen A.S., Foskett J.K. Calcium homeostasis modulator 1 (CALHM1) is the pore-forming subunit of an ion channel that mediates extracellular Ca2+ regulation of neuronal excitability. Proc. Natl. Acad. Sci. USA. 2012;109:E1963–E1971. doi: 10.1073/pnas.1204023109. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

