A Lipoate-Protein Ligase Is Required for De Novo Lipoyl-Protein Biosynthesis in the Hyperthermophilic Archaeon Thermococcus kodakarensis
- PMID: 35736229
- PMCID: PMC9275244
- DOI: 10.1128/aem.00644-22
A Lipoate-Protein Ligase Is Required for De Novo Lipoyl-Protein Biosynthesis in the Hyperthermophilic Archaeon Thermococcus kodakarensis
Abstract
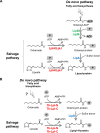
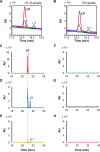
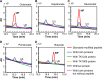
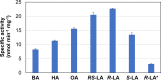
Lipoic acid is an organosulfur cofactor essential for several key enzyme complexes in oxidative and one-carbon metabolism. It is covalently bound to the lipoyl domain of the E2 subunit in some 2-oxoacid dehydrogenase complexes and the H-protein in the glycine cleavage system. Lipoate-protein ligase (Lpl) is involved in the salvage of exogenous lipoate and attaches free lipoate to the E2 subunit or the H-protein in an ATP-dependent manner. In the hyperthermophilic archaeon Thermococcus kodakarensis, TK1234 and TK1908 are predicted to encode the N- and C-terminal regions of Lpl, respectively. TK1908 and TK1234 recombinant proteins form a heterodimer and together displayed significant ligase activity toward octanoate in addition to lipoate when a chemically synthesized octapeptide was used as the acceptor. The proteins also displayed activity toward other fatty acids, indicating broad fatty acid specificity. On the other hand, lipoyl synthase from T. kodakarensis only recognized octanoyl-peptide as a substrate. Examination of individual proteins indicated that the TK1908 protein alone was able to catalyze the ligase reaction although with a much lower activity. Gene disruption of TK1908 led to lipoate/serine auxotrophy, whereas TK1234 gene deletion did not. Acyl carrier protein homologs are not found on the archaeal genomes, and the TK1908/TK1234 protein complex did not utilize octanoyl-CoA, raising the possibility that the substrate of the ligase reaction is octanoic acid itself. Although Lpl has been considered as an enzyme involved in lipoate salvage, the results imply that in T. kodakarensis, the TK1908 and TK1234 proteins function in de novo lipoyl-protein biosynthesis. IMPORTANCE Based on previous studies in bacteria and eukaryotes, lipoate-protein ligases (Lpls) have been considered to be involved exclusively in lipoate salvage. The genetic analyses in this study on the lipoate-protein ligase in T. kodakarensis, however, suggest otherwise and that the enzyme is additionally involved in de novo protein lipoylation. We also provide biochemical evidence that the lipoate-protein ligase displays broad substrate specificity and is capable of ligating acyl groups of various chain-lengths to the peptide substrate. We show that this apparent ambiguity in Lpl is resolved by the strict substrate specificity of the lipoyl synthase LipS in this organism, which only recognizes octanoyl-peptide. The results provide relevant physiological insight into archaeal protein lipoylation.
Keywords: Archaea; biosynthesis; hyperthermophile; lipoate-protein ligase; lipoylation; metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Lipoic acid attachment to proteins: stimulating new developments.Microbiol Mol Biol Rev. 2024 Jun 27;88(2):e0000524. doi: 10.1128/mmbr.00005-24. Epub 2024 Apr 16. Microbiol Mol Biol Rev. 2024. PMID: 38624243 Review.
-
A Structurally Novel Lipoyl Synthase in the Hyperthermophilic Archaeon Thermococcus kodakarensis.Appl Environ Microbiol. 2020 Nov 10;86(23):e01359-20. doi: 10.1128/AEM.01359-20. Print 2020 Nov 10. Appl Environ Microbiol. 2020. PMID: 32978128 Free PMC article.
-
The role of the Saccharomyces cerevisiae lipoate protein ligase homologue, Lip3, in lipoic acid synthesis.Yeast. 2013 Oct;30(10):415-27. doi: 10.1002/yea.2979. Epub 2013 Sep 2. Yeast. 2013. PMID: 23960015 Free PMC article.
-
Identification of Dephospho-Coenzyme A (Dephospho-CoA) Kinase in Thermococcus kodakarensis and Elucidation of the Entire CoA Biosynthesis Pathway in Archaea.mBio. 2019 Jul 23;10(4):e01146-19. doi: 10.1128/mBio.01146-19. mBio. 2019. PMID: 31337720 Free PMC article.
-
Lipoic acid biosynthesis defects.J Inherit Metab Dis. 2014 Jul;37(4):553-63. doi: 10.1007/s10545-014-9705-8. Epub 2014 Apr 29. J Inherit Metab Dis. 2014. PMID: 24777537 Review.
Cited by
-
Removal of phosphoglycolate in hyperthermophilic archaea.Proc Natl Acad Sci U S A. 2024 Apr 16;121(16):e2311390121. doi: 10.1073/pnas.2311390121. Epub 2024 Apr 9. Proc Natl Acad Sci U S A. 2024. PMID: 38593075 Free PMC article.
-
Identification of a novel lipoic acid biosynthesis pathway reveals the complex evolution of lipoate assembly in prokaryotes.PLoS Biol. 2023 Jun 27;21(6):e3002177. doi: 10.1371/journal.pbio.3002177. eCollection 2023 Jun. PLoS Biol. 2023. PMID: 37368881 Free PMC article.
-
Lipoic acid attachment to proteins: stimulating new developments.Microbiol Mol Biol Rev. 2024 Jun 27;88(2):e0000524. doi: 10.1128/mmbr.00005-24. Epub 2024 Apr 16. Microbiol Mol Biol Rev. 2024. PMID: 38624243 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

