PrecisionFDA Truth Challenge V2: Calling variants from short and long reads in difficult-to-map regions
- PMID: 35720974
- PMCID: PMC9205427
- DOI: 10.1016/j.xgen.2022.100129
PrecisionFDA Truth Challenge V2: Calling variants from short and long reads in difficult-to-map regions
Abstract
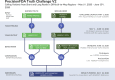
The precisionFDA Truth Challenge V2 aimed to assess the state of the art of variant calling in challenging genomic regions. Starting with FASTQs, 20 challenge participants applied their variant-calling pipelines and submitted 64 variant call sets for one or more sequencing technologies (Illumina, PacBio HiFi, and Oxford Nanopore Technologies). Submissions were evaluated following best practices for benchmarking small variants with updated Genome in a Bottle benchmark sets and genome stratifications. Challenge submissions included numerous innovative methods, with graph-based and machine learning methods scoring best for short-read and long-read datasets, respectively. With machine learning approaches, combining multiple sequencing technologies performed particularly well. Recent developments in sequencing and variant calling have enabled benchmarking variants in challenging genomic regions, paving the way for the identification of previously unknown clinically relevant variants.
Conflict of interest statement
DECLARATION OF INTERESTS C.B. is an employee and shareholder of SAGA Diagnostics AB. A.C., P.-C.C., A.K., M.N., G.B., S.G., and H.Y. are employees of Google, and A.C. is a shareholder. S.D.-B., D.K.-Z., D.T., Ö.K., G.B., K.N., E.A., R.B., I.J.J., A.D., V.S., A.J., and H.S.T. are employees of Seven Bridges Genomics. O.S. and S. T.W. are employees of DNAnexus. G.L., C.M., L.T.F., Y.D., and S.Z. are employees of Genetalks. V.J., M.R., B.L., C.R., S.C., and R.M. are employees of Illumina. S.M.E.S. and M.M. are employees of Roche. C.H. is an employee of Wasai Technology. H.F., Z.L., and L.C. are employees of Sentieon.
Figures





Similar articles
-
Benchmarking datasets for assembly-based variant calling using high-fidelity long reads.BMC Genomics. 2023 Mar 27;24(1):148. doi: 10.1186/s12864-023-09255-y. BMC Genomics. 2023. PMID: 36973656 Free PMC article.
-
Local read haplotagging enables accurate long-read small variant calling.bioRxiv [Preprint]. 2023 Sep 12:2023.09.07.556731. doi: 10.1101/2023.09.07.556731. bioRxiv. 2023. Update in: Nat Commun. 2024 Jul 13;15(1):5907. doi: 10.1038/s41467-024-50079-5. PMID: 37745389 Free PMC article. Updated. Preprint.
-
Benchmarking challenging small variants with linked and long reads.Cell Genom. 2022 May;2(5):100128. doi: 10.1016/j.xgen.2022.100128. Cell Genom. 2022. PMID: 36452119 Free PMC article.
-
Performance analysis of conventional and AI-based variant callers using short and long reads.BMC Bioinformatics. 2023 Dec 14;24(1):472. doi: 10.1186/s12859-023-05596-3. BMC Bioinformatics. 2023. PMID: 38097928 Free PMC article.
-
Variant calling and benchmarking in an era of complete human genome sequences.Nat Rev Genet. 2023 Jul;24(7):464-483. doi: 10.1038/s41576-023-00590-0. Epub 2023 Apr 14. Nat Rev Genet. 2023. PMID: 37059810 Review.
Cited by
-
Comprehensive genome analysis and variant detection at scale using DRAGEN.Nat Biotechnol. 2024 Oct 25. doi: 10.1038/s41587-024-02382-1. Online ahead of print. Nat Biotechnol. 2024. PMID: 39455800
-
The GIAB genomic stratifications resource for human reference genomes.Nat Commun. 2024 Oct 19;15(1):9029. doi: 10.1038/s41467-024-53260-y. Nat Commun. 2024. PMID: 39424793 Free PMC article.
-
Dissecting the Reduced Penetrance of Putative Loss-of-Function Variants in Population-Scale Biobanks.medRxiv [Preprint]. 2024 Oct 7:2024.09.23.24314008. doi: 10.1101/2024.09.23.24314008. medRxiv. 2024. PMID: 39399029 Free PMC article. Preprint.
-
StratoMod: predicting sequencing and variant calling errors with interpretable machine learning.Commun Biol. 2024 Oct 13;7(1):1316. doi: 10.1038/s42003-024-06981-1. Commun Biol. 2024. PMID: 39397114 Free PMC article.
-
Benchmarking reveals superiority of deep learning variant callers on bacterial nanopore sequence data.Elife. 2024 Oct 10;13:RP98300. doi: 10.7554/eLife.98300. Elife. 2024. PMID: 39388235 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

