Context-dependent enhancer function revealed by targeted inter-TAD relocation
- PMID: 35715427
- PMCID: PMC9205857
- DOI: 10.1038/s41467-022-31241-3
Context-dependent enhancer function revealed by targeted inter-TAD relocation
Abstract
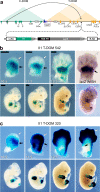
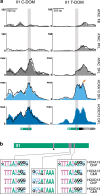
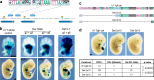
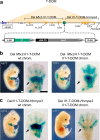
The expression of some genes depends on large, adjacent regions of the genome that contain multiple enhancers. These regulatory landscapes frequently align with Topologically Associating Domains (TADs), where they integrate the function of multiple similar enhancers to produce a global, TAD-specific regulation. We asked if an individual enhancer could overcome the influence of one of these landscapes, to drive gene transcription. To test this, we transferred an enhancer from its native location, into a nearby TAD with a related yet different functional specificity. We used the biphasic regulation of Hoxd genes during limb development as a paradigm. These genes are first activated in proximal limb cells by enhancers located in one TAD, which is then silenced when the neighboring TAD activates its enhancers in distal limb cells. We transferred a distal limb enhancer into the proximal limb TAD and found that its new context suppresses its normal distal specificity, even though it is bound by HOX13 transcription factors, which are responsible for the distal activity. This activity can be rescued only when a large portion of the surrounding environment is removed. These results indicate that, at least in some cases, the functioning of enhancer elements is subordinated to the host chromatin context, which can exert a dominant control over its activity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Impact of genome architecture on the functional activation and repression of Hox regulatory landscapes.BMC Biol. 2019 Jul 12;17(1):55. doi: 10.1186/s12915-019-0677-x. BMC Biol. 2019. PMID: 31299961 Free PMC article.
-
The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes.Genes Dev. 2017 Nov 15;31(22):2264-2281. doi: 10.1101/gad.307769.117. Epub 2017 Dec 22. Genes Dev. 2017. PMID: 29273679 Free PMC article.
-
Chromatin topology and the timing of enhancer function at the HoxD locus.Proc Natl Acad Sci U S A. 2020 Dec 8;117(49):31231-31241. doi: 10.1073/pnas.2015083117. Epub 2020 Nov 23. Proc Natl Acad Sci U S A. 2020. PMID: 33229569 Free PMC article.
-
Are TADs supercoiled?Nucleic Acids Res. 2019 Jan 25;47(2):521-532. doi: 10.1093/nar/gky1091. Nucleic Acids Res. 2019. PMID: 30395328 Free PMC article. Review.
-
Chromatin topology in development and disease.Curr Opin Genet Dev. 2019 Apr;55:32-38. doi: 10.1016/j.gde.2019.04.007. Epub 2019 May 21. Curr Opin Genet Dev. 2019. PMID: 31125724 Review.
Cited by
-
EVOLUTIONARY CO-OPTION OF AN ANCESTRAL CLOACAL REGULATORY LANDSCAPE DURING THE EMERGENCE OF DIGITS AND GENITALS.bioRxiv [Preprint]. 2024 Mar 27:2024.03.24.586442. doi: 10.1101/2024.03.24.586442. bioRxiv. 2024. PMID: 38585989 Free PMC article. Preprint.
-
eQTL mapping in fetal-like pancreatic progenitor cells reveals early developmental insights into diabetes risk.Nat Commun. 2023 Oct 30;14(1):6928. doi: 10.1038/s41467-023-42560-4. Nat Commun. 2023. PMID: 37903777 Free PMC article.
-
Mutations of PDS5 genes enhance TAD-like domain formation Arabidopsis thaliana.Nat Commun. 2024 Oct 29;15(1):9308. doi: 10.1038/s41467-024-53760-x. Nat Commun. 2024. PMID: 39468060 Free PMC article.
-
Function and Evolution of the Loop Extrusion Machinery in Animals.Int J Mol Sci. 2023 Mar 6;24(5):5017. doi: 10.3390/ijms24055017. Int J Mol Sci. 2023. PMID: 36902449 Free PMC article. Review.
-
Three-dimensional genome structure and function.MedComm (2020). 2023 Jul 8;4(4):e326. doi: 10.1002/mco2.326. eCollection 2023 Aug. MedComm (2020). 2023. PMID: 37426677 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

