A Satellite dsRNA Attenuates the Induction of Helper Virus-Mediated Symptoms in Aspergillus flavus
- PMID: 35711767
- PMCID: PMC9195127
- DOI: 10.3389/fmicb.2022.895844
A Satellite dsRNA Attenuates the Induction of Helper Virus-Mediated Symptoms in Aspergillus flavus
Abstract
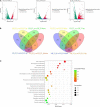
Aspergillus flavus is an important fungal pathogen of animals and plants. Previously, we reported a novel partitivirus, Aspergillus flavus partitivirus 1 (AfPV1), infecting A. flavus. In this study, we obtained a small double-stranded (ds) RNA segment (734 bp), which is a satellite RNA of the helper virus, AfPV1. The presence of AfPV1 altered the colony morphology, decreased the number of conidiophores, created significantly larger vacuoles, and caused more sensitivity to osmotic, oxidative, and UV stresses in A. flavus, but the small RNA segment could attenuate the above symptoms caused by the helper virus AfPV1 in A. flavus. Moreover, AfPV1 infection reduced the pathogenicity of A. flavus in corn (Zea mays), honeycomb moth (Galleria mellonella), mice (Mus musculus), and the adhesion of conidia to host epithelial cells, and increased conidial death by macrophages. However, the small RNA segment could also attenuate the above symptoms caused by the helper virus AfPV1 in A. flavus, perhaps by reducing the genomic accumulation of the helper virus AfPV1 in A. flavus. We used this model to investigate transcriptional genes regulated by AfPV1 and the small RNA segment in A. flavus, and their role in generating different phenotypes. We found that the pathways of the genes regulated by AfPV1 in its host were similar to those of retroviral viruses. Therefore, some pathways may be of benefit to non-retroviral viral integration or endogenization into the genomes of its host. Moreover, some potential antiviral substances were also found in A. flavus using this system.
Keywords: Aspergillus flavus; Aspergillus flavus partitivirus 1; a satellite dsRNA; transcriptional analysis; virus therapy.
Copyright © 2022 Jiang, Yang, Liu, Tian, Wang, Wang, Zhang, Yu, Qi, Jiang and Hsiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




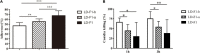
Similar articles
-
Molecular Characterization of a Debilitation-Associated Partitivirus Infecting the Pathogenic Fungus Aspergillus flavus.Front Microbiol. 2019 Mar 28;10:626. doi: 10.3389/fmicb.2019.00626. eCollection 2019. Front Microbiol. 2019. PMID: 30984147 Free PMC article.
-
Melanin depletion affects Aspergillus flavus conidial surface proteins, architecture, and virulence.Appl Microbiol Biotechnol. 2024 Apr 9;108(1):291. doi: 10.1007/s00253-024-13107-4. Appl Microbiol Biotechnol. 2024. PMID: 38592509 Free PMC article.
-
A Network Approach of Gene Co-expression in the Zea mays/Aspergillus flavus Pathosystem to Map Host/Pathogen Interaction Pathways.Front Genet. 2016 Nov 21;7:206. doi: 10.3389/fgene.2016.00206. eCollection 2016. Front Genet. 2016. PMID: 27917194 Free PMC article.
-
Regulation of Conidiogenesis in Aspergillus flavus.Cells. 2022 Sep 7;11(18):2796. doi: 10.3390/cells11182796. Cells. 2022. PMID: 36139369 Free PMC article. Review.
-
Satellite RNA pathogens of plants: impacts and origins-an RNA silencing perspective.Wiley Interdiscip Rev RNA. 2016 Jan-Feb;7(1):5-16. doi: 10.1002/wrna.1311. Epub 2015 Oct 20. Wiley Interdiscip Rev RNA. 2016. PMID: 26481458 Review.
Cited by
-
Identification and Characterization of a Novel Hypovirus from the Phytopathogenic Fungus Botryosphaeria dothidea.Viruses. 2023 Oct 7;15(10):2059. doi: 10.3390/v15102059. Viruses. 2023. PMID: 37896836 Free PMC article.
-
A collection of Trichoderma isolates from natural environments in Sardinia reveals a complex virome that includes negative-sense fungal viruses with unprecedented genome organizations.Virus Evol. 2023 Jul 4;9(2):vead042. doi: 10.1093/ve/vead042. eCollection 2023. Virus Evol. 2023. PMID: 37692893 Free PMC article.
-
The Expanding Mycovirome of Aspergilli.J Fungi (Basel). 2024 Aug 17;10(8):585. doi: 10.3390/jof10080585. J Fungi (Basel). 2024. PMID: 39194910 Free PMC article. Review.
-
RNA interference of Aspergillus flavus in response to Aspergillus flavus partitivirus 1 infection.Front Microbiol. 2023 Nov 14;14:1252294. doi: 10.3389/fmicb.2023.1252294. eCollection 2023. Front Microbiol. 2023. PMID: 38033556 Free PMC article.
-
AfuPmV-1-Infected Aspergillus fumigatus Is More Susceptible to Stress Than Virus-Free Fungus.J Fungi (Basel). 2023 Jul 15;9(7):750. doi: 10.3390/jof9070750. J Fungi (Basel). 2023. PMID: 37504738 Free PMC article.
References
LinkOut - more resources
Full Text Sources

