p166 links membrane and intramitochondrial modules of the trypanosomal tripartite attachment complex
- PMID: 35709300
- PMCID: PMC9242489
- DOI: 10.1371/journal.ppat.1010207
p166 links membrane and intramitochondrial modules of the trypanosomal tripartite attachment complex
Abstract
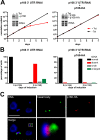
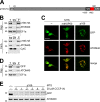
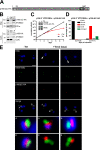
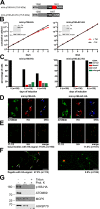
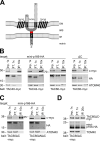
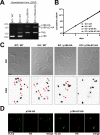
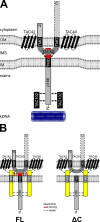
The protist parasite Trypanosoma brucei has a single mitochondrion with a single unit genome termed kinetoplast DNA (kDNA). Faithfull segregation of replicated kDNA is ensured by a complicated structure termed tripartite attachment complex (TAC). The TAC physically links the basal body of the flagellum with the kDNA spanning the two mitochondrial membranes. Here, we characterized p166 as the only known TAC subunit that is anchored in the inner membrane. Its C-terminal transmembrane domain separates the protein into a large N-terminal region that interacts with the kDNA-localized TAC102 and a 34 aa C-tail that binds to the intermembrane space-exposed loop of the integral outer membrane protein TAC60. Whereas the outer membrane region requires four essential subunits for proper TAC function, the inner membrane integral p166, via its interaction with TAC60 and TAC102, would theoretically suffice to bridge the distance between the OM and the kDNA. Surprisingly, non-functional p166 lacking the C-terminal 34 aa still localizes to the TAC region. This suggests the existence of additional TAC-associated proteins which loosely bind to non-functional p166 lacking the C-terminal 34 aa and keep it at the TAC. However, binding of full length p166 to these TAC-associated proteins alone would not be sufficient to withstand the mechanical load imposed by the segregating basal bodies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation.EMBO J. 2008 Jan 9;27(1):143-54. doi: 10.1038/sj.emboj.7601956. Epub 2007 Dec 6. EMBO J. 2008. PMID: 18059470 Free PMC article.
-
TAC102 Is a Novel Component of the Mitochondrial Genome Segregation Machinery in Trypanosomes.PLoS Pathog. 2016 May 11;12(5):e1005586. doi: 10.1371/journal.ppat.1005586. eCollection 2016 May. PLoS Pathog. 2016. PMID: 27168148 Free PMC article.
-
Characterization of the novel mitochondrial genome segregation factor TAP110 in Trypanosoma brucei.J Cell Sci. 2021 Mar 8;134(5):jcs254300. doi: 10.1242/jcs.254300. J Cell Sci. 2021. PMID: 33589495 Free PMC article.
-
Failure is not an option - mitochondrial genome segregation in trypanosomes.J Cell Sci. 2018 Sep 17;131(18):jcs221820. doi: 10.1242/jcs.221820. J Cell Sci. 2018. PMID: 30224426 Review.
-
DNA segregation in mitochondria and beyond: insights from the trypanosomal tripartite attachment complex.Trends Biochem Sci. 2023 Dec;48(12):1058-1070. doi: 10.1016/j.tibs.2023.08.012. Epub 2023 Sep 27. Trends Biochem Sci. 2023. PMID: 37775421 Review.
Cited by
-
Pam16 and Pam18 were repurposed during Trypanosoma brucei evolution to regulate the replication of mitochondrial DNA.PLoS Biol. 2024 Aug 15;22(8):e3002449. doi: 10.1371/journal.pbio.3002449. eCollection 2024 Aug. PLoS Biol. 2024. PMID: 39146359 Free PMC article.
-
Single p197 molecules of the mitochondrial genome segregation system of Trypanosoma brucei determine the distance between basal body and outer membrane.Proc Natl Acad Sci U S A. 2022 Oct 4;119(40):e2204294119. doi: 10.1073/pnas.2204294119. Epub 2022 Sep 26. Proc Natl Acad Sci U S A. 2022. PMID: 36161893 Free PMC article.
-
Characterization of two novel proteins involved in mitochondrial DNA anchoring in Trypanosoma brucei.PLoS Pathog. 2023 Jul 17;19(7):e1011486. doi: 10.1371/journal.ppat.1011486. eCollection 2023 Jul. PLoS Pathog. 2023. PMID: 37459364 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

