Single-Cell Imaging Shows That the Transcriptional State of the HIV-1 Provirus and Its Reactivation Potential Depend on the Integration Site
- PMID: 35708287
- PMCID: PMC9426465
- DOI: 10.1128/mbio.00007-22
Single-Cell Imaging Shows That the Transcriptional State of the HIV-1 Provirus and Its Reactivation Potential Depend on the Integration Site
Abstract
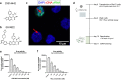
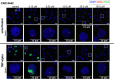
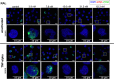
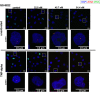
Current antiretroviral treatment fails to cure HIV-1 infection since latent provirus resides in long-lived cellular reservoirs, rebounding whenever therapy is discontinued. The molecular mechanisms underlying HIV-1 latency are complex where the possible link between integration and transcription is poorly understood. HIV-1 integration is targeted toward active chromatin by the direct interaction with a host protein, lens epithelium-derived growth factor (LEDGF/p75). LEDGINs are small-molecule inhibitors of the LEDGF/p75-integrase (IN) interaction that effectively inhibit and retarget HIV-1 integration out of preferred integration sites, resulting in residual provirus that is more latent. Here, we describe a single-cell branched DNA imaging method for simultaneous detection of viral DNA and RNA. We investigated how treatment with LEDGINs affects the location, transcription, and reactivation of HIV-1 in both cell lines and primary cells. This approach demonstrated that LEDGIN-mediated retargeting hampered the baseline transcriptional state and the transcriptional reactivation of the provirus, evidenced by the reduction in viral RNA expression per residual copy. Moreover, treatment of primary cells with LEDGINs induced an enrichment of provirus in deep latency. These results corroborate the impact of integration site selection for the HIV-1 transcriptional state and support block-and-lock functional cure strategies in which the latent reservoir is permanently silenced after retargeting. IMPORTANCE A longstanding question exists on the impact of the HIV-1 integration site on viral gene expression. This unsolved question has significant implications for the search toward an HIV-1 cure, as eradication strategies set up to reactivate and eliminate HIV-1 depend on the site where the provirus is integrated. The main determinant for integration site selection is the interaction of the HIV-1 integrase (IN) and the host chromatin targeting factor, LEDGF/p75. LEDGINs are small-molecule inhibitors of the LEDGF/p75-IN interaction that inhibit and retarget HIV-1 integration out of preferred integration sites. Using both LEDGINs and branched DNA (bDNA) imaging, we now investigated, in much detail, the impact of integration site selection on the three-dimensional location of the provirus, HIV-1 transcription, and reactivation. Our results provide evidence for a "block-and-lock" functional cure strategy that aims to permanently silence HIV-1 by LEDGIN-mediated retargeting to sites that are less susceptible to reactivation after treatment interruption.
Keywords: HIV-1; LEDGF/p75; LEDGINs; integration site selection; single-cell HIV-1 imaging.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Insight in HIV Integration Site Selection Provides a Block-and-Lock Strategy for a Functional Cure of HIV Infection.Viruses. 2018 Dec 26;11(1):12. doi: 10.3390/v11010012. Viruses. 2018. PMID: 30587760 Free PMC article. Review.
-
Impact of LEDGIN treatment during virus production on residual HIV-1 transcription.Retrovirology. 2019 Apr 2;16(1):8. doi: 10.1186/s12977-019-0472-3. Retrovirology. 2019. PMID: 30940165 Free PMC article.
-
LEDGIN-mediated Inhibition of Integrase-LEDGF/p75 Interaction Reduces Reactivation of Residual Latent HIV.EBioMedicine. 2016 Jun;8:248-264. doi: 10.1016/j.ebiom.2016.04.039. Epub 2016 May 13. EBioMedicine. 2016. PMID: 27428435 Free PMC article.
-
The chromatin landscape at the HIV-1 provirus integration site determines viral expression.Nucleic Acids Res. 2020 Aug 20;48(14):7801-7817. doi: 10.1093/nar/gkaa536. Nucleic Acids Res. 2020. PMID: 32597987 Free PMC article.
-
Towards a Functional Cure of HIV-1: Insight Into the Chromatin Landscape of the Provirus.Front Microbiol. 2021 Mar 18;12:636642. doi: 10.3389/fmicb.2021.636642. eCollection 2021. Front Microbiol. 2021. PMID: 33868195 Free PMC article. Review.
Cited by
-
Interventions during Early Infection: Opening a Window for an HIV Cure?Viruses. 2024 Oct 9;16(10):1588. doi: 10.3390/v16101588. Viruses. 2024. PMID: 39459922 Free PMC article. Review.
-
Mechanisms and efficacy of small molecule latency-promoting agents to inhibit HIV reactivation ex vivo.JCI Insight. 2024 Aug 20;9(19):e183084. doi: 10.1172/jci.insight.183084. JCI Insight. 2024. PMID: 39163135 Free PMC article.
-
Specific quantification of inducible HIV-1 reservoir by RT-LAMP.Commun Med (Lond). 2024 Jun 25;4(1):123. doi: 10.1038/s43856-024-00553-4. Commun Med (Lond). 2024. PMID: 38918506 Free PMC article.
-
SIV infection and ARV treatment reshape the transcriptional and epigenetic profile of naïve and memory T cells in vivo.J Virol. 2024 Jun 13;98(6):e0028324. doi: 10.1128/jvi.00283-24. Epub 2024 May 23. J Virol. 2024. PMID: 38780248 Free PMC article.
-
Viral and host mediators of non-suppressible HIV-1 viremia.Nat Med. 2023 Dec;29(12):3212-3223. doi: 10.1038/s41591-023-02611-1. Epub 2023 Nov 13. Nat Med. 2023. PMID: 37957382 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

