Single-molecule study of the effects of temperature, pH, and RNA base on the stepwise enzyme kinetics of 10-23 deoxyribozyme
- PMID: 35702195
- PMCID: PMC9113834
- DOI: 10.1039/d2ra02131e
Single-molecule study of the effects of temperature, pH, and RNA base on the stepwise enzyme kinetics of 10-23 deoxyribozyme
Abstract
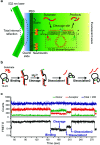
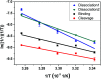
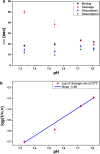
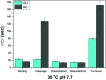
We investigated how the stepwise enzyme kinetics of 10-23 deoxyribozyme was affected by temperature, pH, and RNA residue of the substrate at the single-molecule level. A deoxyribozyme-substrate system was employed to temporally categorize a single-turnover reaction into four distinct steps: binding, cleavage, dissociation of one of the cleaved fragments, and dissociation of the other fragment. The dwell time of each step was measured as the temperature was varied from 26 to 34 °C, to which the transition state theory was applied to obtain the enthalpy and entropy of activation for individual steps. In addition, we found that only the cleavage step was significantly affected by pH, indicating that it involves deprotonation of a single proton. We also found that different RNA residues specifically affect the cleavage step and cause the dwell time to change by as much as 5 times.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures
Similar articles
-
A Novel Small RNA-Cleaving Deoxyribozyme with a Short Binding Arm.Sci Rep. 2019 Jun 3;9(1):8224. doi: 10.1038/s41598-019-44750-x. Sci Rep. 2019. PMID: 31160698 Free PMC article.
-
Characterization of a local folding event of the Tetrahymena group I ribozyme: effects of oligonucleotide substrate length, pH, and temperature on the two substrate binding steps.Biochemistry. 1999 Oct 26;38(43):14192-204. doi: 10.1021/bi9914309. Biochemistry. 1999. PMID: 10571993
-
Kinetic and thermodynamic characterization of the RNA-cleaving 8-17 deoxyribozyme.Nucleic Acids Res. 2004 Feb 12;32(3):916-25. doi: 10.1093/nar/gkh250. Print 2004. Nucleic Acids Res. 2004. PMID: 14963261 Free PMC article.
-
Effects of metal ions and catalytic loop sequences on the complex formation of a deoxyribozyme and its RNA substrate.J Inorg Biochem. 2000 Nov;82(1-4):189-95. doi: 10.1016/s0162-0134(00)00159-8. J Inorg Biochem. 2000. PMID: 11132626
-
Kinetic scheme for intermolecular RNA cleavage by a ribozyme derived from hepatitis delta virus RNA.Biochemistry. 2000 Aug 8;39(31):9055-66. doi: 10.1021/bi000499+. Biochemistry. 2000. PMID: 10924098
References
-
- Payen A. Persoz J. F. Ann. Chim. Phys. 1833;53:73.
LinkOut - more resources
Full Text Sources

