Discovery of Histone Deacetylase Inhibitor Using Molecular Modeling and Free Energy Calculations
- PMID: 35694501
- PMCID: PMC9178742
- DOI: 10.1021/acsomega.2c01572
Discovery of Histone Deacetylase Inhibitor Using Molecular Modeling and Free Energy Calculations
Abstract
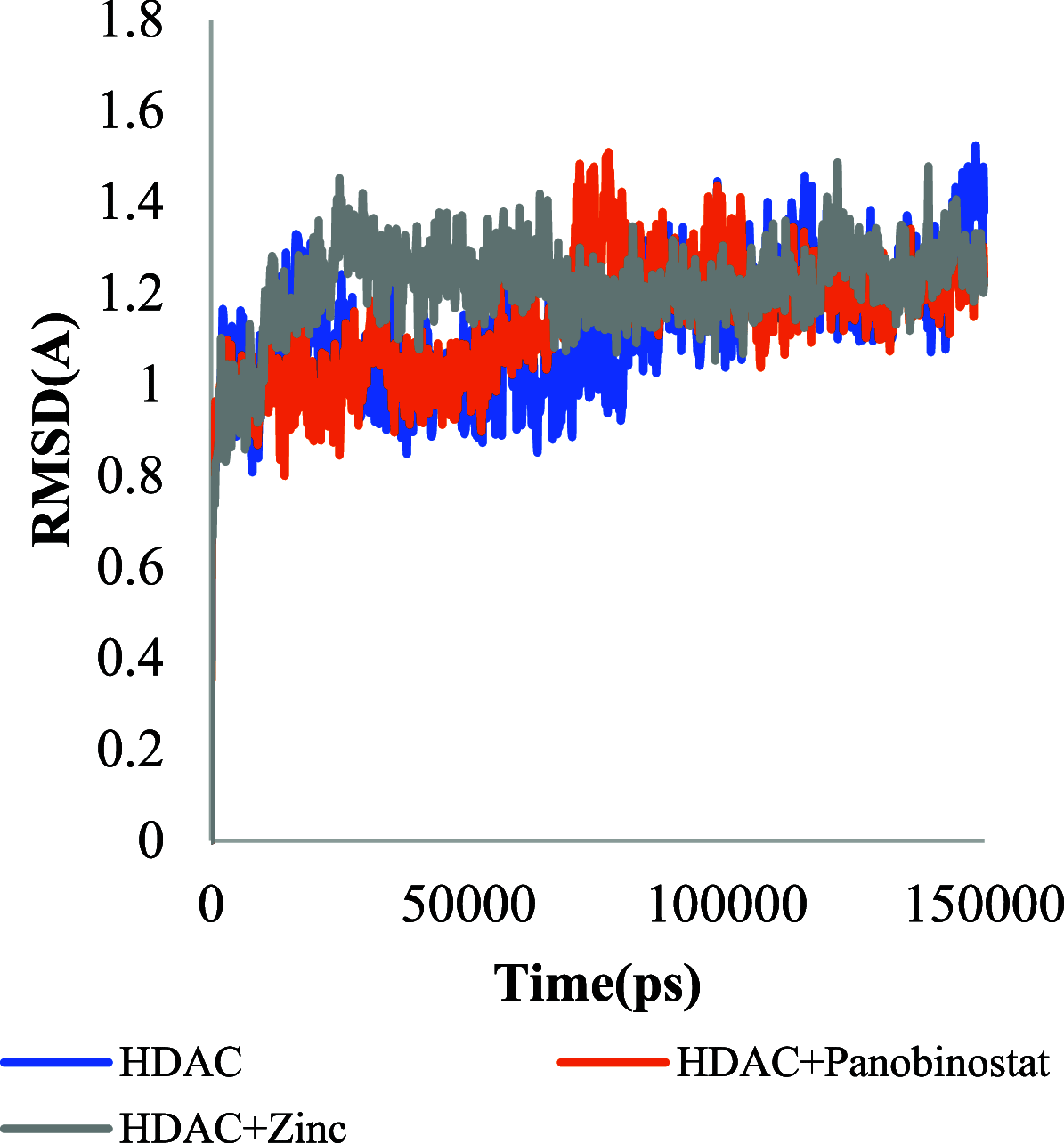
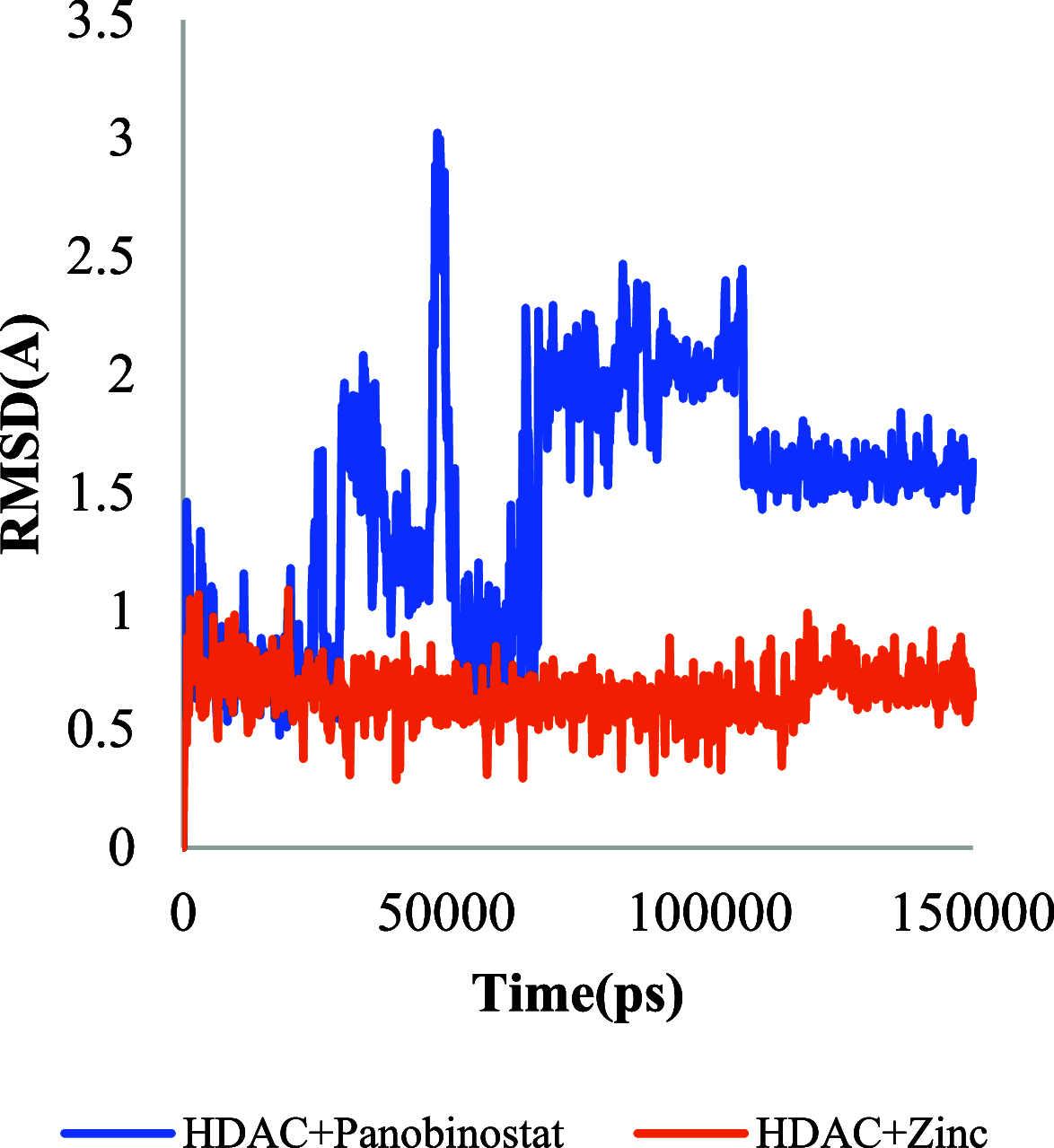
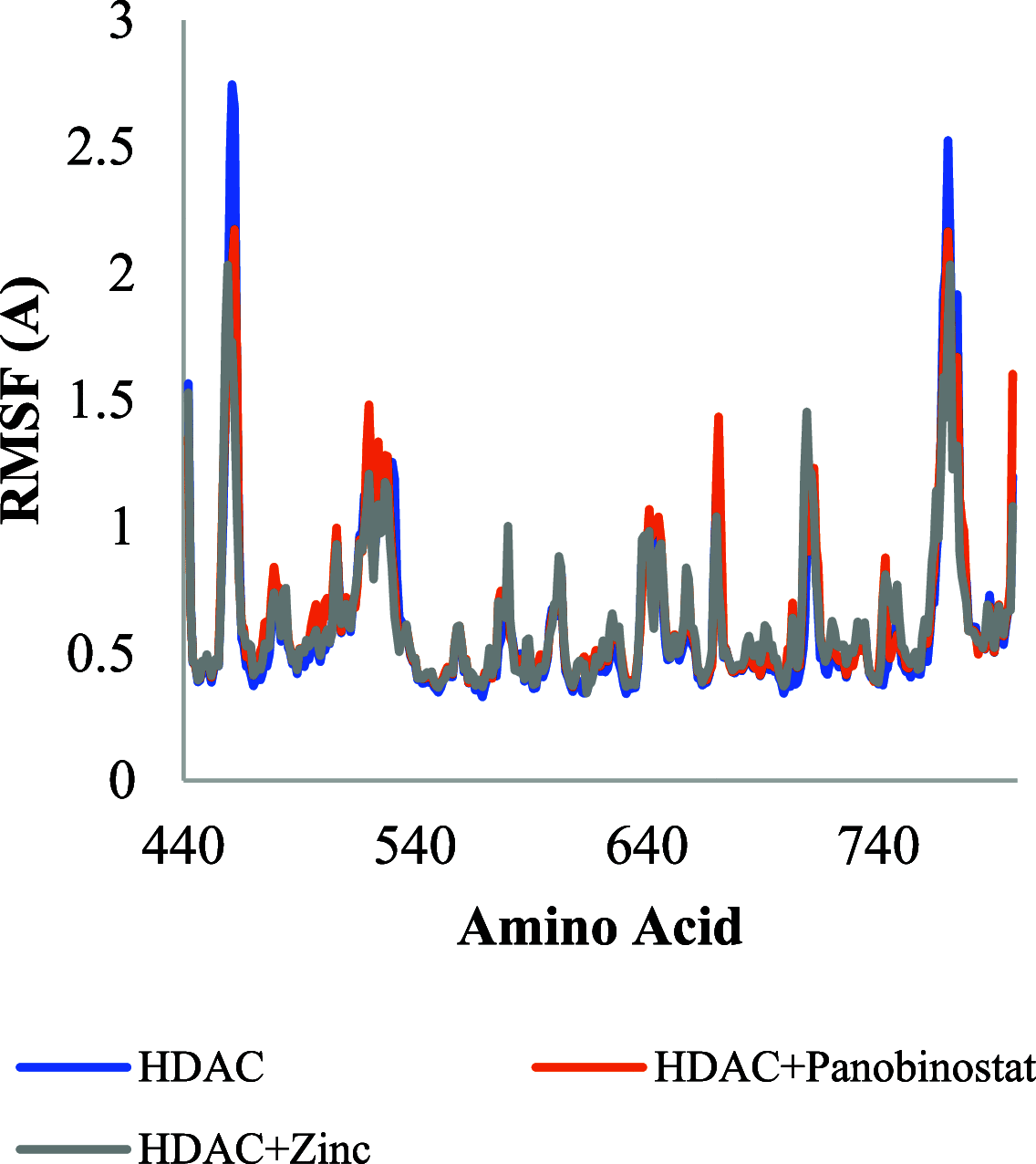
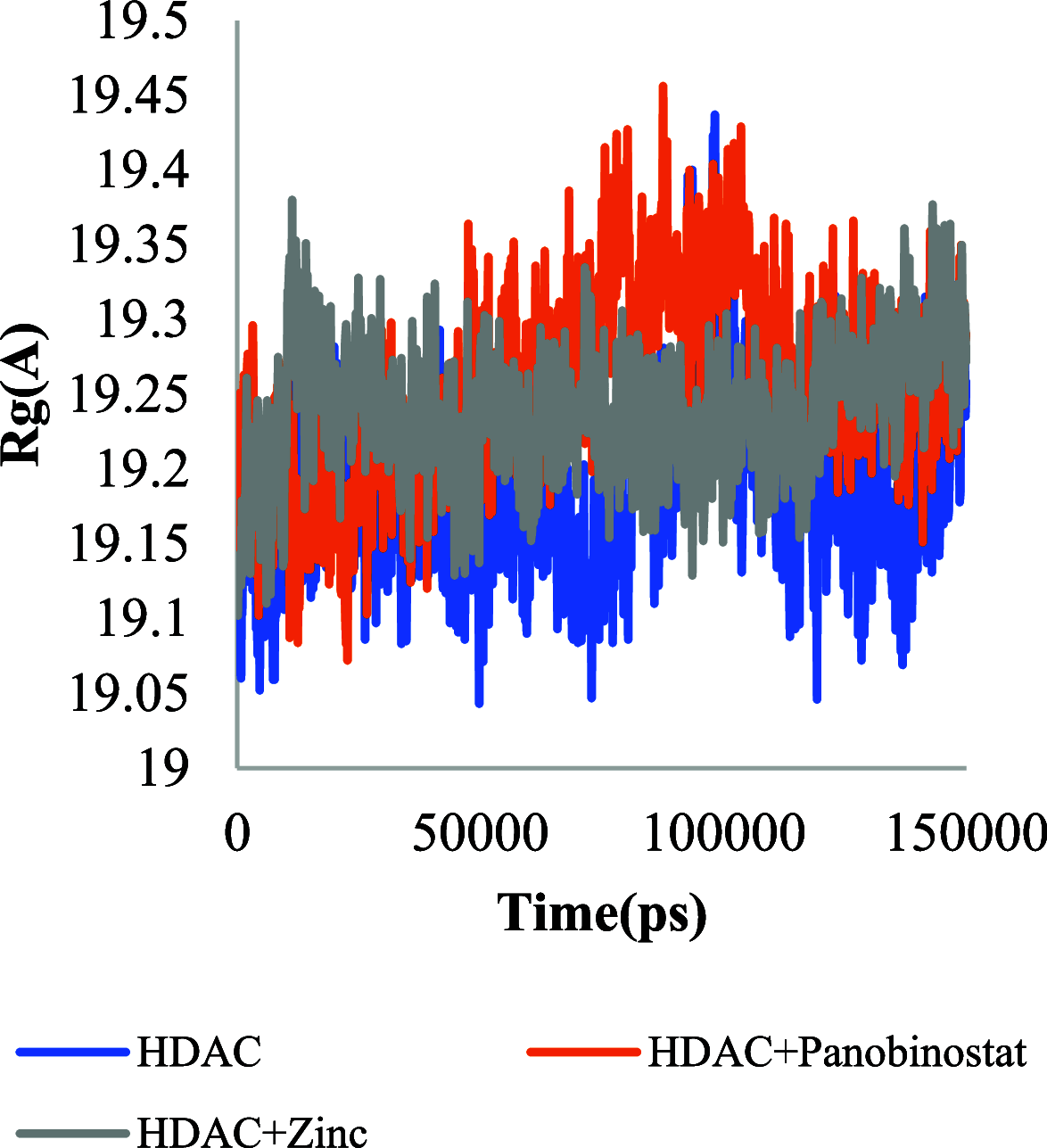
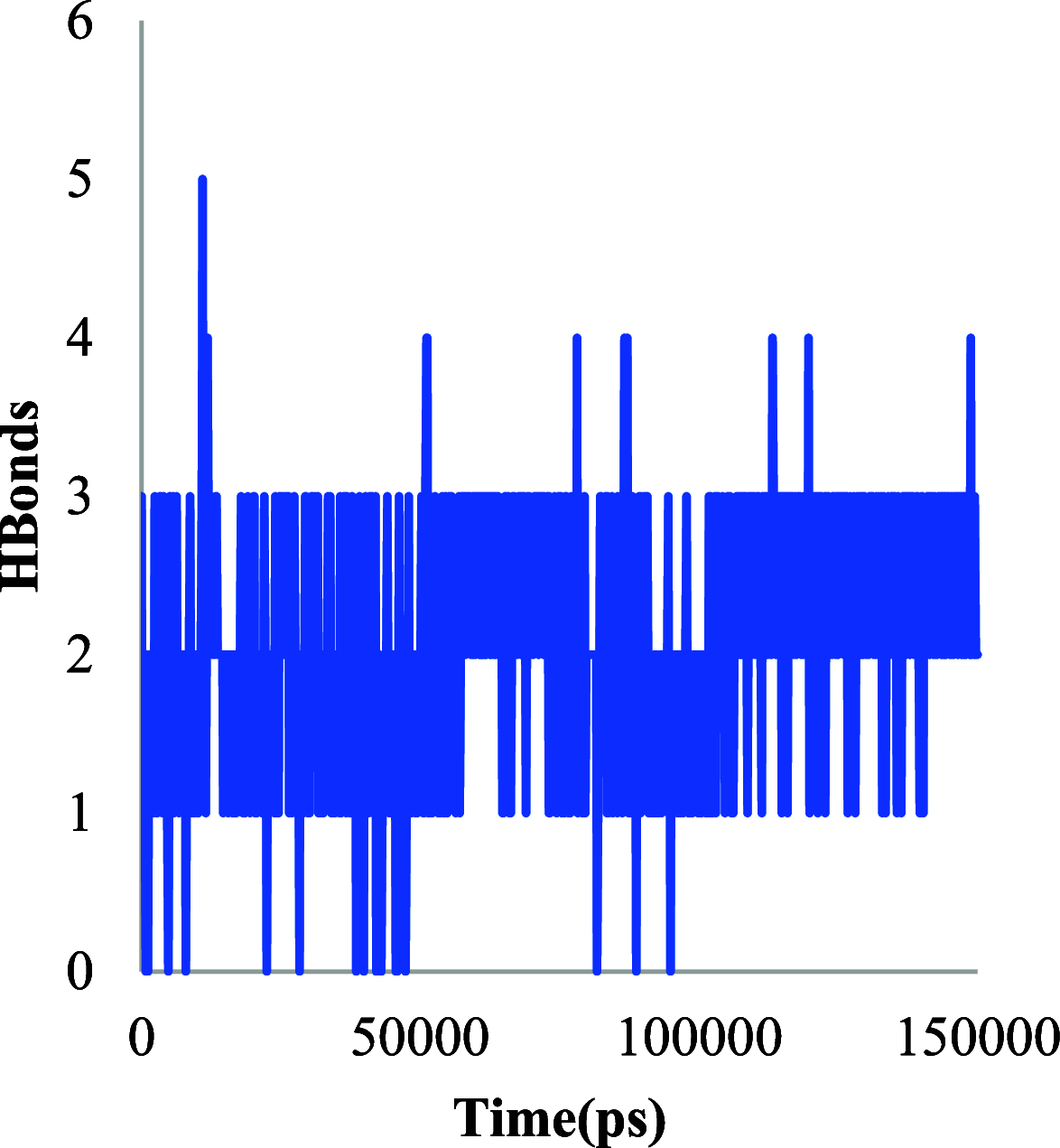
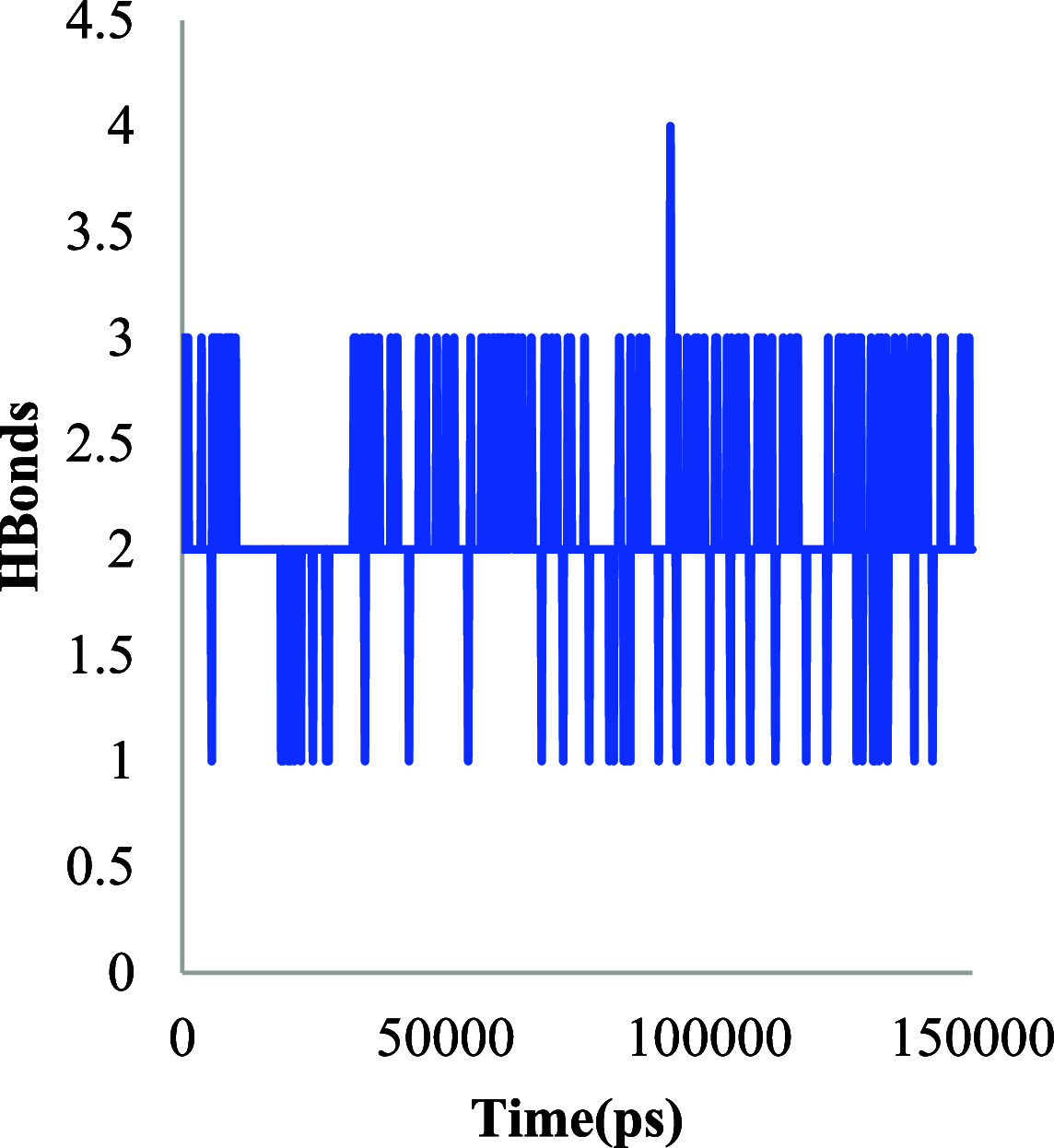
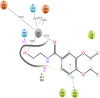
The histone acetylation-deacetylation at lysine regulates the functions of many cellular proteins. An increased expression of HDAC6 can cause an increased amount of deacetylated histones, which leads to an inhibition of gene expression and has been associated with cancer cell proliferation. The present study screened the ZINC database to find novel HDAC6 inhibitors using virtual high-throughput screening techniques. The docking score, free energy, and binding pattern of the complexes were used to select a best ligand for further study. Molecular dynamic simulations, binding interactions, and the stability of docked conformations were investigated. Several parameters that determine protein-ligand interactions, such as root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), radius of gyration (Rg), and binding pattern, were observed. Hydrogen bonds were observed at His 573 and Gly 582 after a 150 ns simulation with identified compound ZINC000002845205, and they were similar to known inhibitor Panobinostat. The molecular mechanics with generalised Born and surface area solvation (MM/GBSA) free energy was comparable to known inhibitor Panobinostat. ZINC000002845205 qualifies drug-likeness according to Lipinski's rule-of-five, rule-of-three, and the World Drug Index (WDI)-like rule, but there is one violation in the lead-like rule.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
Discovery of the allosteric inhibitor from actinomyces metabolites to target EGFRCSTMLR mutant protein: molecular modeling and free energy approach.Sci Rep. 2023 Jun 1;13(1):8885. doi: 10.1038/s41598-023-33065-7. Sci Rep. 2023. PMID: 37264083 Free PMC article.
-
Molecular modelling study to discover novel JAK2 signaling pathway inhibitor.J Biomol Struct Dyn. 2023 Jul-Aug;41(12):5827-5838. doi: 10.1080/07391102.2022.2097314. Epub 2022 Jul 15. J Biomol Struct Dyn. 2023. PMID: 35838147
-
Investigation of non-hydroxamate scaffolds against HDAC6 inhibition: A pharmacophore modeling, molecular docking, and molecular dynamics simulation approach.J Bioinform Comput Biol. 2018 Jun;16(3):1840015. doi: 10.1142/S0219720018400152. J Bioinform Comput Biol. 2018. PMID: 29945500
-
Identification of anti-filarial leads against aspartate semialdehyde dehydrogenase of Wolbachia endosymbiont of Brugia malayi: combined molecular docking and molecular dynamics approaches.J Biomol Struct Dyn. 2019 Feb;37(2):394-410. doi: 10.1080/07391102.2018.1427633. Epub 2018 Feb 6. J Biomol Struct Dyn. 2019. PMID: 29334340
-
Posttranslational modification and beyond: interplay between histone deacetylase 6 and heat-shock protein 90.Mol Med. 2021 Sep 16;27(1):110. doi: 10.1186/s10020-021-00375-3. Mol Med. 2021. PMID: 34530730 Free PMC article. Review.
Cited by
-
Discovery of the allosteric inhibitor from actinomyces metabolites to target EGFRCSTMLR mutant protein: molecular modeling and free energy approach.Sci Rep. 2023 Jun 1;13(1):8885. doi: 10.1038/s41598-023-33065-7. Sci Rep. 2023. PMID: 37264083 Free PMC article.
-
Novel hydroxamic acid derivative induces apoptosis and constrains autophagy in leukemic cells.J Adv Res. 2024 Jun;60:201-214. doi: 10.1016/j.jare.2023.07.005. Epub 2023 Jul 17. J Adv Res. 2024. PMID: 37467961 Free PMC article.
-
Indole-containing pharmaceuticals: targets, pharmacological activities, and SAR studies.RSC Med Chem. 2024 Jan 30;15(3):788-808. doi: 10.1039/d3md00677h. eCollection 2024 Mar 20. RSC Med Chem. 2024. PMID: 38516587 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

