Coming full circle: On the origin and evolution of the looping model for enhancer-promoter communication
- PMID: 35691341
- PMCID: PMC9283939
- DOI: 10.1016/j.jbc.2022.102117
Coming full circle: On the origin and evolution of the looping model for enhancer-promoter communication
Abstract
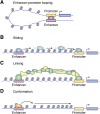
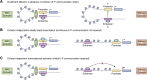
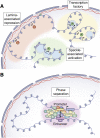
In mammalian organisms, enhancers can regulate transcription from great genomic distances. How enhancers affect distal gene expression has been a major question in the field of gene regulation. One model to explain how enhancers communicate with their target promoters, the chromatin looping model, posits that enhancers and promoters come in close spatial proximity to mediate communication. Chromatin looping has been broadly accepted as a means for enhancer-promoter communication, driven by accumulating in vitro and in vivo evidence. The genome is now known to be folded into a complex 3D arrangement, created and maintained in part by the interplay of the Cohesin complex and the DNA-binding protein CTCF. In the last few years, however, doubt over the relationship between looping and transcriptional activation has emerged, driven by studies finding that only a modest number of genes are perturbed with acute degradation of looping machinery components. In parallel, newer models describing distal enhancer action have also come to prominence. In this article, we explore the emergence and development of the looping model as a means for enhancer-promoter communication and review the contrasting evidence between historical gene-specific and current global data for the role of chromatin looping in transcriptional regulation. We also discuss evidence for alternative models to chromatin looping and their support in the literature. We suggest that, while there is abundant evidence for chromatin looping as a major mechanism for enhancer function, enhancer-promoter communication is likely mediated by more than one mechanism in an enhancer- and context-dependent manner.
Keywords: 3D genome; CTCF; Cohesin; TAD; chromatin structure; genome structure; promoter; transcription; transcription enhancer.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Similar articles
-
Impact of 3D genome organization, guided by cohesin and CTCF looping, on sex-biased chromatin interactions and gene expression in mouse liver.Epigenetics Chromatin. 2020 Jul 17;13(1):30. doi: 10.1186/s13072-020-00350-y. Epigenetics Chromatin. 2020. PMID: 32680543 Free PMC article.
-
Three-dimensional genome architectural CCCTC-binding factor makes choice in duplicated enhancers at Pcdhα locus.Sci China Life Sci. 2020 Jun;63(6):835-844. doi: 10.1007/s11427-019-1598-4. Epub 2020 Apr 2. Sci China Life Sci. 2020. PMID: 32249388
-
ZNF143 deletion alters enhancer/promoter looping and CTCF/cohesin geometry.Cell Rep. 2024 Jan 23;43(1):113663. doi: 10.1016/j.celrep.2023.113663. Epub 2024 Jan 10. Cell Rep. 2024. PMID: 38206813
-
The structural and functional roles of CTCF in the regulation of cell type-specific and human disease-associated super-enhancers.Genes Genomics. 2019 Mar;41(3):257-265. doi: 10.1007/s13258-018-0768-z. Epub 2018 Nov 19. Genes Genomics. 2019. PMID: 30456521 Review.
-
CTCF shapes chromatin structure and gene expression in health and disease.EMBO Rep. 2022 Sep 5;23(9):e55146. doi: 10.15252/embr.202255146. Epub 2022 Aug 22. EMBO Rep. 2022. PMID: 35993175 Free PMC article. Review.
Cited by
-
Cohesin mutations in acute myeloid leukemia.Leukemia. 2024 Nov;38(11):2318-2328. doi: 10.1038/s41375-024-02406-4. Epub 2024 Sep 9. Leukemia. 2024. PMID: 39251741 Review.
-
Next-generation forward genetic screens: uniting high-throughput perturbations with single-cell analysis.Trends Genet. 2024 Feb;40(2):118-133. doi: 10.1016/j.tig.2023.10.012. Epub 2023 Nov 20. Trends Genet. 2024. PMID: 37989654 Review.
-
Through the lens of phase separation: intrinsically unstructured protein and chromatin looping.Nucleus. 2023 Dec;14(1):2179766. doi: 10.1080/19491034.2023.2179766. Nucleus. 2023. PMID: 36821650 Free PMC article. Review.
-
You shall not pass! Unveiling the barriers for cohesin-mediated loop extrusion.Mol Cell. 2022 Jul 21;82(14):2541-2543. doi: 10.1016/j.molcel.2022.06.028. Mol Cell. 2022. PMID: 35868255 Free PMC article.
-
CRISPR tiling deletion screens reveal functional enhancers of neuropsychiatric risk genes and allelic compensation effects (ACE) on transcription.bioRxiv [Preprint]. 2024 Oct 10:2024.10.08.616922. doi: 10.1101/2024.10.08.616922. bioRxiv. 2024. PMID: 39416108 Free PMC article. Preprint.
References
-
- Gillies S.D., Morrison S.L., Oi V.T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–728. - PubMed
-
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. - PubMed
-
- Mercola M., Wang X.F., Olsen J., Calame K. Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus. Science. 1983;221:663–665. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

