Exploring the Effect of Mechanical Anisotropy of Protein Structures in the Unfoldase Mechanism of AAA+ Molecular Machines
- PMID: 35683705
- PMCID: PMC9182431
- DOI: 10.3390/nano12111849
Exploring the Effect of Mechanical Anisotropy of Protein Structures in the Unfoldase Mechanism of AAA+ Molecular Machines
Abstract
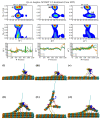
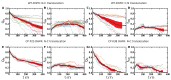
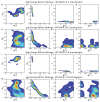
Essential cellular processes of microtubule disassembly and protein degradation, which span lengths from tens of μm to nm, are mediated by specialized molecular machines with similar hexameric structure and function. Our molecular simulations at atomistic and coarse-grained scales show that both the microtubule-severing protein spastin and the caseinolytic protease ClpY, accomplish spectacular unfolding of their diverse substrates, a microtubule lattice and dihydrofolate reductase (DHFR), by taking advantage of mechanical anisotropy in these proteins. Unfolding of wild-type DHFR requires disruption of mechanically strong β-sheet interfaces near each terminal, which yields branched pathways associated with unzipping along soft directions and shearing along strong directions. By contrast, unfolding of circular permutant DHFR variants involves single pathways due to softer mechanical interfaces near terminals, but translocation hindrance can arise from mechanical resistance of partially unfolded intermediates stabilized by β-sheets. For spastin, optimal severing action initiated by pulling on a tubulin subunit is achieved through specific orientation of the machine versus the substrate (microtubule lattice). Moreover, changes in the strength of the interactions between spastin and a microtubule filament, which can be driven by the tubulin code, lead to drastically different outcomes for the integrity of the hexameric structure of the machine.
Keywords: AAA+ superfamily; allostery; microtubule severing; molecular dynamics; molecular machines; protein degradation.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




Similar articles
-
Molecular investigations into the unfoldase action of severing enzymes on microtubules.Cytoskeleton (Hoboken). 2020 May;77(5-6):214-228. doi: 10.1002/cm.21606. Epub 2020 Mar 25. Cytoskeleton (Hoboken). 2020. PMID: 32170815
-
Asymmetric Conformational Transitions in AAA+ Biological Nanomachines Modulate Direction-Dependent Substrate Protein Unfolding Mechanisms.J Phys Chem B. 2017 Jul 27;121(29):7108-7121. doi: 10.1021/acs.jpcb.7b05963. Epub 2017 Jul 18. J Phys Chem B. 2017. PMID: 28675036
-
Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin.Nature. 2008 Jan 17;451(7176):363-7. doi: 10.1038/nature06482. Nature. 2008. PMID: 18202664 Free PMC article.
-
Meiotic Clade AAA ATPases: Protein Polymer Disassembly Machines.J Mol Biol. 2016 May 8;428(9 Pt B):1897-911. doi: 10.1016/j.jmb.2015.11.004. Epub 2015 Nov 10. J Mol Biol. 2016. PMID: 26555750 Free PMC article. Review.
-
[Microtubule severing proteins - structure and activity regulation].Postepy Biochem. 2016;62(1):46-51. Postepy Biochem. 2016. PMID: 28132444 Review. Polish.
Cited by
-
Protein Nanomechanics.Nanomaterials (Basel). 2022 Oct 8;12(19):3524. doi: 10.3390/nano12193524. Nanomaterials (Basel). 2022. PMID: 36234652 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

