Characterization of the Interaction Between SARS-CoV-2 Membrane Protein (M) and Proliferating Cell Nuclear Antigen (PCNA) as a Potential Therapeutic Target
- PMID: 35677658
- PMCID: PMC9168989
- DOI: 10.3389/fcimb.2022.849017
Characterization of the Interaction Between SARS-CoV-2 Membrane Protein (M) and Proliferating Cell Nuclear Antigen (PCNA) as a Potential Therapeutic Target
Abstract
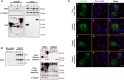
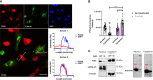
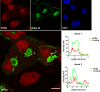
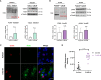
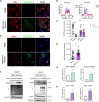
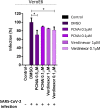
SARS-CoV-2 is an emerging virus from the Coronaviridae family and is responsible for the ongoing COVID-19 pandemic. In this work, we explored the previously reported SARS-CoV-2 structural membrane protein (M) interaction with human Proliferating Cell Nuclear Antigen (PCNA). The M protein is responsible for maintaining virion shape, and PCNA is a marker of DNA damage which is essential for DNA replication and repair. We validated the M-PCNA interaction through immunoprecipitation, immunofluorescence co-localization, and PLA (Proximity Ligation Assay). In cells infected with SARS-CoV-2 or transfected with M protein, using immunofluorescence and cell fractioning, we documented a reallocation of PCNA from the nucleus to the cytoplasm and the increase of PCNA and γH2AX (another DNA damage marker) expression. We also observed an increase in PCNA and γH2AX expression in the lung of a COVID-19 patient by immunohistochemistry. In addition, the inhibition of PCNA translocation by PCNA I1 and Verdinexor led to a reduction of plaque formation in an in vitro assay. We, therefore, propose that the transport of PCNA to the cytoplasm and its association with M could be a virus strategy to manipulate cell functions and may be considered a target for COVID-19 therapy.
Keywords: DNA damage; M protein; PCNA; SARS-CoV-2; viral-host interaction.
Copyright © 2022 Zambalde, Pavan, Mancini, Severino, Scudero, Morelli, Amorim, Bispo-dos-Santos, Góis, Toledo-Teixeira, Parise, Mauad, Dolhnikoff, Saldiva, Marques-Souza, Proenca-Modena, Ventura and Simabuco.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Ubiquitin‑like protein FAT10 regulates DNA damage repair via modification of proliferating cell nuclear antigen.Mol Med Rep. 2018 Jun;17(6):7487-7496. doi: 10.3892/mmr.2018.8843. Epub 2018 Apr 5. Mol Med Rep. 2018. PMID: 29620277 Free PMC article.
-
The herpes simplex virus 1 UL36USP deubiquitinase suppresses DNA repair in host cells via deubiquitination of proliferating cell nuclear antigen.J Biol Chem. 2017 May 19;292(20):8472-8483. doi: 10.1074/jbc.M117.778076. Epub 2017 Mar 27. J Biol Chem. 2017. PMID: 28348081 Free PMC article.
-
Nuclear insulin-like growth factor 1 receptor phosphorylates proliferating cell nuclear antigen and rescues stalled replication forks after DNA damage.J Biol Chem. 2017 Nov 3;292(44):18227-18239. doi: 10.1074/jbc.M117.781492. Epub 2017 Sep 18. J Biol Chem. 2017. PMID: 28924044 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
The Many Roles of PCNA in Eukaryotic DNA Replication.Enzymes. 2016;39:231-54. doi: 10.1016/bs.enz.2016.03.003. Epub 2016 Apr 19. Enzymes. 2016. PMID: 27241932 Free PMC article. Review.
Cited by
-
Immune characteristics of kidney transplant recipients with acute respiratory distress syndrome induced by COVID-19 at single-cell resolution.Respir Res. 2024 Jan 18;25(1):34. doi: 10.1186/s12931-024-02682-9. Respir Res. 2024. PMID: 38238762 Free PMC article.
-
Zebrafish models of COVID-19.FEMS Microbiol Rev. 2023 Jan 16;47(1):fuac042. doi: 10.1093/femsre/fuac042. FEMS Microbiol Rev. 2023. PMID: 36323404 Free PMC article. Review.
-
Computational Analysis of Short Linear Motifs in the Spike Protein of SARS-CoV-2 Variants Provides Possible Clues into the Immune Hijack and Evasion Mechanisms of Omicron Variant.Int J Mol Sci. 2022 Aug 8;23(15):8822. doi: 10.3390/ijms23158822. Int J Mol Sci. 2022. PMID: 35955954 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

