Extracellular sialyltransferase st6gal1 in breast tumor cell growth and invasiveness
- PMID: 35676533
- PMCID: PMC9663294
- DOI: 10.1038/s41417-022-00485-y
Extracellular sialyltransferase st6gal1 in breast tumor cell growth and invasiveness
Abstract
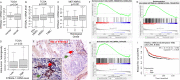
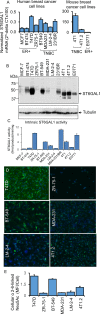
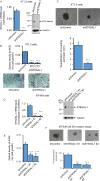
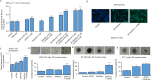
The sialyltransferase ST6GAL1 that adds α2-6 linked sialic acids to N-glycans of cell surface and secreted glycoproteins is prominently associated with many human cancers. Tumor-native ST6GAL1 promotes tumor cell behaviors such as invasion and resistance to cell stress and chemo- and radio-treatments. Canonically, ST6GAL1 resides in the intracellular secretory apparatus and glycosylates nascent glycoproteins in biosynthetic transit. However, ST6GAL1 is also released into the extracellular milieu and extracellularly remodels cell surface and secreted glycans. The impact of this non-canonical extrinsic mechanism of ST6GAL1 on tumor cell pathobiology is not known. We hypothesize that ST6GAL1 action is the combined effect of natively expressed sialyltransferase acting cell-autonomously within the ER-Golgi complex and sialyltransferase from extracellular origins acting extrinsically to remodel cell-surface glycans. We found that shRNA knockdown of intrinsic ST6GAL1 expression resulted in decreased ST6GAL1 cargo in the exosome-like vesicles as well as decreased breast tumor cell growth and invasive behavior in 3D in vitro cultures. Extracellular ST6GAL1, present in cancer exosomes or the freely soluble recombinant sialyltransferase, compensates for insufficient intrinsic ST6GAL1 by boosting cancer cell proliferation and increasing invasiveness. Moreover, we present evidence supporting the existence novel but yet uncharacterized cofactors in the exosome-like particles that potently amplify extrinsic ST6GAL1 action, highlighting a previously unknown mechanism linking this enzyme and cancer pathobiology. Our data indicate that extracellular ST6GAL1 from remote sources can compensate for cellular ST6GAL1-mediated aggressive tumor cell proliferation and invasive behavior and has great clinical potential for extracellular ST6GAL1 as these molecules are in the extracellular space should be easily accessible targets.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Regulation of ST6GAL1 sialyltransferase expression in cancer cells.Glycobiology. 2021 Jun 3;31(5):530-539. doi: 10.1093/glycob/cwaa110. Glycobiology. 2021. PMID: 33320246 Free PMC article. Review.
-
ST6GAL1-mediated aberrant sialylation promotes prostate cancer progression.J Pathol. 2023 Sep;261(1):71-84. doi: 10.1002/path.6152. Epub 2023 Aug 7. J Pathol. 2023. PMID: 37550801
-
Galectin-1-mediated cell adhesion, invasion and cell death in human anaplastic large cell lymphoma: regulatory roles of cell surface glycans.Int J Oncol. 2014 May;44(5):1433-42. doi: 10.3892/ijo.2014.2319. Epub 2014 Mar 4. Int J Oncol. 2014. PMID: 24589677 Free PMC article.
-
Majority of alpha2,6-sialylated glycans in the adult mouse brain exist in O-glycans: SALSA-MS analysis for knockout mice of alpha2,6-sialyltransferase genes.Glycobiology. 2021 Jun 3;31(5):557-570. doi: 10.1093/glycob/cwaa105. Glycobiology. 2021. PMID: 33242079
-
ST6GAL1: A key player in cancer.Oncol Lett. 2019 Aug;18(2):983-989. doi: 10.3892/ol.2019.10458. Epub 2019 Jun 7. Oncol Lett. 2019. PMID: 31423157 Free PMC article. Review.
Cited by
-
Cancer-Derived Extracellular Vesicles as Biomarkers for Cutaneous Squamous Cell Carcinoma: A Systematic Review.Cancers (Basel). 2022 Oct 18;14(20):5098. doi: 10.3390/cancers14205098. Cancers (Basel). 2022. PMID: 36291882 Free PMC article. Review.
-
Glycosphingolipids: from metabolism to chemoenzymatic total synthesis.Org Biomol Chem. 2024 Aug 22;22(33):6665-6683. doi: 10.1039/d4ob00695j. Org Biomol Chem. 2024. PMID: 39120686 Free PMC article. Review.
-
Intratumoral Influenza Vaccine Administration Attenuates Breast Cancer Growth and Restructures the Tumor Microenvironment through Sialic Acid Binding of Vaccine Hemagglutinin.Int J Mol Sci. 2023 Dec 22;25(1):225. doi: 10.3390/ijms25010225. Int J Mol Sci. 2023. PMID: 38203396 Free PMC article.
-
Aberrant Sialylation in Cancer: Therapeutic Opportunities.Cancers (Basel). 2022 Aug 31;14(17):4248. doi: 10.3390/cancers14174248. Cancers (Basel). 2022. PMID: 36077781 Free PMC article. Review.
-
Sialyltransferases and Neuraminidases: Potential Targets for Cancer Treatment.Diseases. 2022 Nov 28;10(4):114. doi: 10.3390/diseases10040114. Diseases. 2022. PMID: 36547200 Free PMC article. Review.
References
-
- Swindall AF, Londono-Joshi AI, Schultz MJ, Fineberg N, Buchsbaum DJ, Bellis SL. ST6Gal-I protein expression is upregulated in human epithelial tumors and correlates with stem cell markers in normal tissues and colon cancer cell lines. Cancer Res. 2013;73:2368–78. doi: 10.1158/0008-5472.CAN-12-3424. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

