Studying the Dynamics of a Complex G-Quadruplex System: Insights into the Comparison of MD and NMR Data
- PMID: 35666124
- PMCID: PMC9281369
- DOI: 10.1021/acs.jctc.2c00291
Studying the Dynamics of a Complex G-Quadruplex System: Insights into the Comparison of MD and NMR Data
Abstract
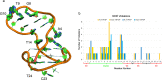
Molecular dynamics (MD) simulations are coming of age in the study of nucleic acids, including specific tertiary structures such as G-quadruplexes. While being precious for providing structural and dynamic information inaccessible to experiments at the atomistic level of resolution, MD simulations in this field may still be limited by several factors. These include the force fields used, different models for ion parameters, ionic strengths, and water models. We address various aspects of this problem by analyzing and comparing microsecond-long atomistic simulations of the G-quadruplex structure formed by the human immunodeficiency virus long terminal repeat (HIV LTR)-III sequence for which nuclear magnetic resonance (NMR) structures are available. The system is studied in different conditions, systematically varying the ionic strengths, ion numbers, and water models. We comparatively analyze the dynamic behavior of the G-quadruplex motif in various conditions and assess the ability of each simulation to satisfy the nuclear magnetic resonance (NMR)-derived experimental constraints and structural parameters. The conditions taking into account K+-ions to neutralize the system charge, mimicking the intracellular ionic strength, and using the four-atom water model are found to be the best in reproducing the experimental NMR constraints and data. Our analysis also reveals that in all of the simulated environments residues belonging to the duplex moiety of HIV LTR-III exhibit the highest flexibility.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Molecular Dynamics Simulation Study of Parallel Telomeric DNA Quadruplexes at Different Ionic Strengths: Evaluation of Water and Ion Models.J Phys Chem B. 2016 Aug 4;120(30):7380-91. doi: 10.1021/acs.jpcb.6b06485. Epub 2016 Jul 26. J Phys Chem B. 2016. PMID: 27379924
-
Direct NMR detection of alkali metal ions bound to G-quadruplex DNA.J Am Chem Soc. 2008 Mar 19;130(11):3590-602. doi: 10.1021/ja709975z. Epub 2008 Feb 23. J Am Chem Soc. 2008. PMID: 18293981
-
Computational and Experimental Characterization of rDNA and rRNA G-Quadruplexes.J Phys Chem B. 2022 Jan 27;126(3):609-619. doi: 10.1021/acs.jpcb.1c08340. Epub 2022 Jan 13. J Phys Chem B. 2022. PMID: 35026949 Free PMC article.
-
NMR spectroscopy of G-quadruplexes.Methods. 2012 May;57(1):11-24. doi: 10.1016/j.ymeth.2012.05.003. Epub 2012 May 24. Methods. 2012. PMID: 22633887 Review.
-
G-quadruplexes and metal ions.Met Ions Life Sci. 2012;10:119-34. doi: 10.1007/978-94-007-2172-2_4. Met Ions Life Sci. 2012. PMID: 22210337 Review.
Cited by
-
From RNA sequence to its three-dimensional structure: geometrical structure, stability and dynamics of selected fragments of SARS-CoV-2 RNA.NAR Genom Bioinform. 2024 Jun 4;6(2):lqae062. doi: 10.1093/nargab/lqae062. eCollection 2024 Jun. NAR Genom Bioinform. 2024. PMID: 38835951 Free PMC article.
-
Computational Modeling of DNA 3D Structures: From Dynamics and Mechanics to Folding.Molecules. 2023 Jun 17;28(12):4833. doi: 10.3390/molecules28124833. Molecules. 2023. PMID: 37375388 Free PMC article. Review.
-
Base pair dynamics, electrostatics, and thermodynamics at the LTR-III quadruplex:duplex junction.Biophys J. 2024 May 7;123(9):1129-1138. doi: 10.1016/j.bpj.2024.03.042. Epub 2024 Apr 4. Biophys J. 2024. PMID: 38576161
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

