Extracellular matrix dynamics: tracking in biological systems and their implications
- PMID: 35637526
- PMCID: PMC9153193
- DOI: 10.1186/s13036-022-00292-x
Extracellular matrix dynamics: tracking in biological systems and their implications
Abstract
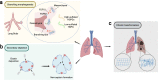
The extracellular matrix (ECM) constitutes the main acellular microenvironment of cells in almost all tissues and organs. The ECM not only provides mechanical support, but also mediates numerous biochemical interactions to guide cell survival, proliferation, differentiation, and migration. Thus, better understanding the everchanging temporal and spatial shifts in ECM composition and structure - the ECM dynamics - will provide fundamental insight regarding extracellular regulation of tissue homeostasis and how tissue states transition from one to another during diverse pathophysiological processes. This review outlines the mechanisms mediating ECM-cell interactions and highlights how changes in the ECM modulate tissue development and disease progression, using the lung as the primary model organ. We then discuss existing methodologies for revealing ECM compositional dynamics, with a particular focus on tracking newly synthesized ECM proteins. Finally, we discuss the ramifications ECM dynamics have on tissue engineering and how to implement spatial and temporal specific extracellular microenvironments into bioengineered tissues. Overall, this review communicates the current capabilities for studying native ECM dynamics and delineates new research directions in discovering and implementing ECM dynamics to push the frontier forward.
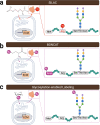
Keywords: Bioorthogonal non-canonical amino acid tagging (BONCAT); Bioprinting; Extracellular matrix (ECM); Lung; Newly synthesized protein; Proteomics; Stable isotope labeling by amino acids in cell culture (SILAC).
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interest.
Figures

Similar articles
-
Targeted proteomics effectively quantifies differences between native lung and detergent-decellularized lung extracellular matrices.Acta Biomater. 2016 Dec;46:91-100. doi: 10.1016/j.actbio.2016.09.043. Epub 2016 Sep 29. Acta Biomater. 2016. PMID: 27693690 Free PMC article.
-
The matrix in focus: new directions in extracellular matrix research from the 2021 ASMB hybrid meeting.Biol Open. 2022 Jan 15;11(1):bio059156. doi: 10.1242/bio.059156. Epub 2022 Jan 7. Biol Open. 2022. PMID: 34994383 Free PMC article.
-
Defining the extracellular matrix using proteomics.Int J Exp Pathol. 2013 Apr;94(2):75-92. doi: 10.1111/iep.12011. Epub 2013 Feb 19. Int J Exp Pathol. 2013. PMID: 23419153 Free PMC article. Review.
-
Recent Trends in Decellularized Extracellular Matrix Bioinks for 3D Printing: An Updated Review.Int J Mol Sci. 2019 Sep 18;20(18):4628. doi: 10.3390/ijms20184628. Int J Mol Sci. 2019. PMID: 31540457 Free PMC article. Review.
-
Engineering Breast Cancer Microenvironments and 3D Bioprinting.Front Bioeng Biotechnol. 2018 May 24;6:66. doi: 10.3389/fbioe.2018.00066. eCollection 2018. Front Bioeng Biotechnol. 2018. PMID: 29881724 Free PMC article. Review.
Cited by
-
The Effect of Substrate Properties on Cellular Behavior and Nanoparticle Uptake in Human Fibroblasts and Epithelial Cells.Nanomaterials (Basel). 2024 Feb 10;14(4):342. doi: 10.3390/nano14040342. Nanomaterials (Basel). 2024. PMID: 38392715 Free PMC article.
-
Trends in extracellular matrix biology.Mol Biol Rep. 2023 Jan;50(1):853-863. doi: 10.1007/s11033-022-07931-y. Epub 2022 Nov 7. Mol Biol Rep. 2023. PMID: 36342580 Free PMC article. Review.
-
Role of Matrix Metalloproteinases in Musculoskeletal Diseases.Biomedicines. 2022 Oct 4;10(10):2477. doi: 10.3390/biomedicines10102477. Biomedicines. 2022. PMID: 36289739 Free PMC article. Review.
-
Extracellular Matrix Proteomics: The mdx-4cv Mouse Diaphragm as a Surrogate for Studying Myofibrosis in Dystrophinopathy.Biomolecules. 2023 Jul 12;13(7):1108. doi: 10.3390/biom13071108. Biomolecules. 2023. PMID: 37509144 Free PMC article. Review.
-
Enhancing tendon-bone integration and healing with advanced multi-layer nanofiber-reinforced 3D scaffolds for acellular tendon complexes.Mater Today Bio. 2024 May 22;26:101099. doi: 10.1016/j.mtbio.2024.101099. eCollection 2024 Jun. Mater Today Bio. 2024. PMID: 38840797 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

