Targeted Inhibition of O-Linked β-N-Acetylglucosamine Transferase as a Promising Therapeutic Strategy to Restore Chemosensitivity and Attenuate Aggressive Tumor Traits in Chemoresistant Urothelial Carcinoma of the Bladder
- PMID: 35625898
- PMCID: PMC9138654
- DOI: 10.3390/biomedicines10051162
Targeted Inhibition of O-Linked β-N-Acetylglucosamine Transferase as a Promising Therapeutic Strategy to Restore Chemosensitivity and Attenuate Aggressive Tumor Traits in Chemoresistant Urothelial Carcinoma of the Bladder
Abstract
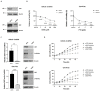
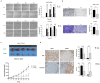
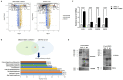
Acquisition of acquired chemoresistance during treatment cycles in urothelial carcinoma of the bladder (UCB) is the major cause of death through enhancing the risk of cancer progression and metastasis. Elevated glucose flux through the abnormal upregulation of O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) controls key signaling and metabolic pathways regulating diverse cancer cell phenotypes. This study showed that OGT expression levels in two human UCB cell models with acquired resistance to gemcitabine and paclitaxel were significantly upregulated compared with those in parental cells. Reducing hyper-O-GlcNAcylation by OGT knockdown (KD) markedly facilitated chemosensitivity to the corresponding chemotherapeutics in both cells, and combination treatment with OGT-KD showed more severe growth defects in chemoresistant sublines. We subsequently verified the suppressive effects of OGT-KD monotherapy on cell migration/invasion in vitro and xenograft tumor growth in vivo in chemoresistant UCB cells. Transcriptome analysis of these cells revealed 97 upregulated genes, which were enriched in multiple oncogenic pathways. Our final choice of suspected OGT glycosylation substrate was VCAN, S1PR3, PDGFRB, and PRKCG, the knockdown of which induced cell growth defects. These findings demonstrate the vital role of dysregulated OGT activity and hyper-O-GlcNAcylation in modulating treatment failure and tumor aggression in chemoresistant UCB.
Keywords: O-linked N-acetylglucosaminylation; O-linked β-N-acetylglucosamine transferase; biomarkers; chemoresistance; gemcitabine; paclitaxel; urothelial carcinoma of bladder.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Suppressed OGT expression inhibits cell proliferation while inducing cell apoptosis in bladder cancer.BMC Cancer. 2018 Nov 20;18(1):1141. doi: 10.1186/s12885-018-5033-y. BMC Cancer. 2018. PMID: 30453909 Free PMC article.
-
Critical role of O-Linked β-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis.J Biol Chem. 2012 Mar 30;287(14):11070-81. doi: 10.1074/jbc.M111.302547. Epub 2012 Jan 24. J Biol Chem. 2012. PMID: 22275356 Free PMC article.
-
Downregulation of O-linked N-acetylglucosamine transferase by RNA interference decreases MMP9 expression in human esophageal cancer cells.Oncol Lett. 2016 May;11(5):3317-3323. doi: 10.3892/ol.2016.4428. Epub 2016 Apr 7. Oncol Lett. 2016. PMID: 27123109 Free PMC article.
-
Essential role of O-GlcNAcylation in stabilization of oncogenic factors.Biochim Biophys Acta Gen Subj. 2019 Aug;1863(8):1302-1317. doi: 10.1016/j.bbagen.2019.04.002. Epub 2019 Apr 26. Biochim Biophys Acta Gen Subj. 2019. PMID: 31034911 Review.
-
OGT as potential novel target: Structure, function and inhibitors.Chem Biol Interact. 2022 Apr 25;357:109886. doi: 10.1016/j.cbi.2022.109886. Epub 2022 Mar 11. Chem Biol Interact. 2022. PMID: 35288161 Review.
Cited by
-
The roles of OGT and its mechanisms in cancer.Cell Biosci. 2024 Sep 16;14(1):121. doi: 10.1186/s13578-024-01301-w. Cell Biosci. 2024. PMID: 39285476 Free PMC article. Review.
-
Altered Glycosylation in Progression and Management of Bladder Cancer.Molecules. 2023 Apr 13;28(8):3436. doi: 10.3390/molecules28083436. Molecules. 2023. PMID: 37110670 Free PMC article. Review.
-
Identification and verification of anoikis-related gene markers to predict the prognosis of patients with bladder cancer and assist in the diagnosis and treatment of bladder cancer.Transl Cancer Res. 2024 Feb 29;13(2):579-593. doi: 10.21037/tcr-23-1770. Epub 2024 Feb 19. Transl Cancer Res. 2024. PMID: 38482431 Free PMC article.
References
-
- Li R., Li Y., Song J., Gao K., Chen K., Yang X., Ding Y., Ma X., Wang Y., Li W., et al. Intravesical gemcitabine versus mitomycin for non-muscle invasive bladder cancer: A systematic review and meta-analysis of randomized controlled trial. BMC Urol. 2020;20:97. doi: 10.1186/s12894-020-00610-9. - DOI - PMC - PubMed
-
- Lu J.L., Xia Q.D., Lu Y.H., Liu Z., Zhou P., Hu H.L., Wang S.G. Efficacy of intravesical therapies on the prevention of recurrence and progression of non-muscle-invasive bladder cancer: A systematic review and network meta-analysis. Cancer Med. 2020;9:7800–7809. doi: 10.1002/cam4.3513. - DOI - PMC - PubMed
-
- Hurle R., Contieri R., Casale P., Morenghi E., Saita A., Buffi N., Lughezzani G., Colombo P., Frego N., Fasulo V., et al. Midterm follow-up (3 years) confirms and extends short-term results of intravesical gemcitabine as bladder-preserving treatment for non-muscle-invasive bladder cancer after BCG failure. Urol. Oncol. 2021;39:195.e7–195.e113. doi: 10.1016/j.urolonc.2020.09.017. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

