Iguratimod Attenuates Macrophage Polarization and Antibody-Mediated Rejection After Renal Transplant by Regulating KLF4
- PMID: 35614941
- PMCID: PMC9125033
- DOI: 10.3389/fphar.2022.865363
Iguratimod Attenuates Macrophage Polarization and Antibody-Mediated Rejection After Renal Transplant by Regulating KLF4
Abstract
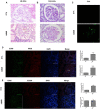
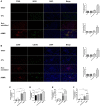
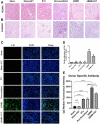
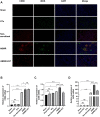
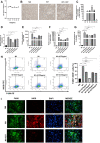
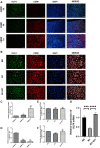
Background: This study aimed to explore the effect and mechanism of iguratimod (IGT) on M1 macrophage polarization and antibody-mediated rejection (ABMR) after renal transplant. Methods: Bioinformatics analysis was performed using three public databases derived from the GEO database. Sprague-Dawley (SD) rats were pre-sensitized with donors of Wistar rats in skin transplantation and a rat renal transplant ABMR model was established from the donors to skin pre-sensitized recipients. Subsequently, IGT was treated on the ABMR model. Routine staining and immunofluorescence (IF) staining were performed to observe the pathological changes in each group and flow cytometry was performed to detect the changes of DSA titers in peripheral blood. In addition, bone-marrow-derived macrophage (BMDM) was extracted and interfered with IGT to explore the effect of IGT in vivo. PCR, IF staining, and Western blot were used to detect the expression of related genes and proteins. Results: Bioinformatics analysis revealed that several immune cells were significantly infiltrated in the ABMR allograft, while M1 macrophage was noticed with the most significance. Results of IF staining and PCR proved the findings of the bioinformatics analysis. Based on this, IGT was observed to significantly attenuate the degree of peritubular capillary vasculitis and arteriolitis in the rat renal transplant ABMR model, whereas it decreases the expression of C4d and reduces the titer of DSA. Results in vitro suggested that M1 macrophage-related transcripts and proteins were significantly reduced by the treatment of IGT in a dose- and time-dependent manner. Furthermore, IGT intervention could remarkably decrease the expression of KLF4. Conclusion: Polarization of M1 macrophages may aggravate ABMR after renal transplant by promoting DSA-mediated endothelial cell injury, and IGT may attenuate the pathogenesis of ABMR by targeting KLF4.
Keywords: KLF4; antibody-mediated rejection; iguratimod; kidney transplant; macrophage polarization.
Copyright © 2022 Hang, Wei, Zheng, Gui, Chen, Sun, Fei, Han, Tao, Wang, Tan and Gu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Iguratimod ameliorates antibody-mediated rejection after renal transplant by modulating the Th17/Treg paradigm.Int Immunopharmacol. 2024 Jul 30;136:112409. doi: 10.1016/j.intimp.2024.112409. Epub 2024 Jun 7. Int Immunopharmacol. 2024. PMID: 38850789
-
Molecular Assessment of C4d-Positive Renal Transplant Biopsies Without Evidence of Rejection.Kidney Int Rep. 2018 Sep 18;4(1):148-158. doi: 10.1016/j.ekir.2018.09.005. eCollection 2019 Jan. Kidney Int Rep. 2018. PMID: 30596178 Free PMC article.
-
Significance of C4d deposition in antibody-mediated rejection.Clin Transplant. 2012 Jul;26 Suppl 24:43-8. doi: 10.1111/j.1399-0012.2012.01642.x. Clin Transplant. 2012. PMID: 22747475
-
Donor-specific antibodies in kidney transplantation: the University of Wisconsin experience.Curr Opin Organ Transplant. 2020 Dec;25(6):543-548. doi: 10.1097/MOT.0000000000000814. Curr Opin Organ Transplant. 2020. PMID: 33044350 Review.
-
Endothelial transcripts uncover a previously unknown phenotype: C4d-negative antibody-mediated rejection.Curr Opin Organ Transplant. 2010 Feb;15(1):42-8. doi: 10.1097/MOT.0b013e3283352a50. Curr Opin Organ Transplant. 2010. PMID: 20009933 Review.
Cited by
-
Iguratimod prevents renal fibrosis in unilateral ureteral obstruction model mice by suppressing M2 macrophage infiltration and macrophage-myofibroblast transition.Ren Fail. 2024 Dec;46(1):2327498. doi: 10.1080/0886022X.2024.2327498. Epub 2024 Apr 26. Ren Fail. 2024. PMID: 38666363 Free PMC article.
-
Macrophage polarization induces endothelium-to-myofibroblast transition in chronic allograft dysfunction.Ren Fail. 2023 Dec;45(1):2220418. doi: 10.1080/0886022X.2023.2220418. Ren Fail. 2023. PMID: 37288756 Free PMC article.
References
-
- Almatroodi S. A., Almatroudi A., Alsahli M. A., Aljasir M. A., Syed M. A., Rahmani A. H. (2020). Epigallocatechin-3-Gallate (EGCG), an Active Compound of Green Tea Attenuates Acute Lung Injury Regulating Macrophage Polarization and Krüpple-Like-Factor 4 (KLF4) Expression. Molecules 25 (12). 10.3390/molecules25122853 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

