Touch signaling and thigmomorphogenesis are regulated by complementary CAMTA3- and JA-dependent pathways
- PMID: 35594358
- PMCID: PMC9122320
- DOI: 10.1126/sciadv.abm2091
Touch signaling and thigmomorphogenesis are regulated by complementary CAMTA3- and JA-dependent pathways
Abstract
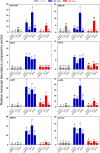
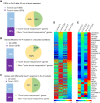
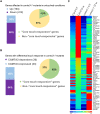
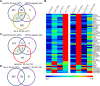
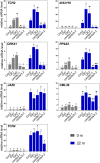
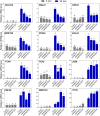
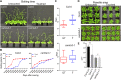
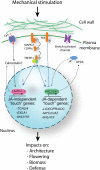
Plants respond to mechanical stimuli to direct their growth and counteract environmental threats. Mechanical stimulation triggers rapid gene expression changes and affects plant appearance (thigmomorphogenesis) and flowering. Previous studies reported the importance of jasmonic acid (JA) in touch signaling. Here, we used reverse genetics to further characterize the molecular mechanisms underlying touch signaling. We show that Piezo mechanosensitive ion channels have no major role in touch-induced gene expression and thigmomorphogenesis. In contrast, the receptor-like kinase Feronia acts as a strong negative regulator of the JA-dependent branch of touch signaling. Last, we show that calmodulin-binding transcriptional activators CAMTA1/2/3 are key regulators of JA-independent touch signaling. CAMTA1/2/3 cooperate to directly bind the promoters and activate gene expression of JA-independent touch marker genes like TCH2 and TCH4. In agreement, camta3 mutants show a near complete loss of thigmomorphogenesis and touch-induced delay of flowering. In conclusion, we have now identified key regulators of two independent touch-signaling pathways.
Figures

Similar articles
-
Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests.Curr Biol. 2012 Apr 24;22(8):701-6. doi: 10.1016/j.cub.2012.02.061. Epub 2012 Apr 5. Curr Biol. 2012. PMID: 22483939
-
A Labor-saving and Repeatable Touch-force Signaling Mutant Screen Protocol for the Study of Thigmomorphogenesis of a Model Plant Arabidopsis thaliana.J Vis Exp. 2019 Aug 6;(150). doi: 10.3791/59392. J Vis Exp. 2019. PMID: 31449235
-
Quantitative and functional posttranslational modification proteomics reveals that TREPH1 plays a role in plant touch-delayed bolting.Proc Natl Acad Sci U S A. 2018 Oct 23;115(43):E10265-E10274. doi: 10.1073/pnas.1814006115. Epub 2018 Oct 5. Proc Natl Acad Sci U S A. 2018. PMID: 30291188 Free PMC article.
-
Mechanical stress acclimation in plants: Linking hormones and somatic memory to thigmomorphogenesis.Plant Cell Environ. 2022 Apr;45(4):989-1010. doi: 10.1111/pce.14252. Epub 2022 Jan 18. Plant Cell Environ. 2022. PMID: 34984703 Review.
-
Thigmomorphogenesis: a complex plant response to mechano-stimulation.J Exp Bot. 2009;60(1):43-56. doi: 10.1093/jxb/ern315. Epub 2008 Dec 16. J Exp Bot. 2009. PMID: 19088336 Review.
Cited by
-
DNA-Binding Activity of CAMTA3 Is Essential for Its Function: Identification of Critical Amino Acids for Its Transcriptional Activity.Cells. 2023 Aug 2;12(15):1986. doi: 10.3390/cells12151986. Cells. 2023. PMID: 37566065 Free PMC article.
-
Mechanosensing, from forces to structures.Front Plant Sci. 2022 Dec 1;13:1060018. doi: 10.3389/fpls.2022.1060018. eCollection 2022. Front Plant Sci. 2022. PMID: 36531357 Free PMC article. Review.
-
Characterizing the role of PP2A B'' family subunits in mechanical stress response and plant development through calcium and ABA signaling in Arabidopsis thaliana.PLoS One. 2024 Nov 14;19(11):e0313590. doi: 10.1371/journal.pone.0313590. eCollection 2024. PLoS One. 2024. PMID: 39541304 Free PMC article.
-
Touch me not! Jasmonic acid and ethylene converge on gibberellins breakdown to regulate touch-induced morphogenesis.Plant Physiol. 2024 Jan 31;194(2):601-603. doi: 10.1093/plphys/kiad588. Plant Physiol. 2024. PMID: 37925742 Free PMC article. No abstract available.
-
FERONIA functions through Target of Rapamycin (TOR) to negatively regulate autophagy.Front Plant Sci. 2022 Aug 23;13:961096. doi: 10.3389/fpls.2022.961096. eCollection 2022. Front Plant Sci. 2022. PMID: 36082288 Free PMC article.
References
-
- Braam J., In touch: Plant responses to mechanical stimuli. New Phytol. 165, 373–389 (2005). - PubMed
-
- Zhou L. H., Liu S. B., Wang P. F., Lu T. J., Xu F., Genin G. M., Pickard B. G., The Arabidopsis trichome is an active mechanosensory switch. Plant Cell Environ. 40, 611–621 (2017). - PubMed
-
- Jaffe M. J., Thigmomorphogenesis: The response of plant growth and development to mechanical stimulation: With special reference to Bryonia dioica. Planta 114, 143–157 (1973). - PubMed
-
- Si T., Wang X., Huang M., Cai J., Zhou Q., Dai T., Jiang D., Double benefits of mechanical wounding in enhancing chilling tolerance and lodging resistance in wheat plants. Plant Biol. (Stuttg.) 21, 813–824 (2019). - PubMed
-
- Tateno M., Increase in lodging safety factor of thigmomorphogenically dwarfed shoots of mulberry tree. Physiol. Plant. 81, 239–243 (1991).
LinkOut - more resources
Full Text Sources

