Revisiting the concept of peptide bond planarity in an iron-sulfur protein by neutron structure analysis
- PMID: 35594350
- PMCID: PMC9122329
- DOI: 10.1126/sciadv.abn2276
Revisiting the concept of peptide bond planarity in an iron-sulfur protein by neutron structure analysis
Abstract
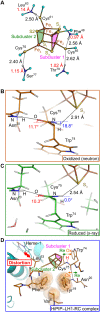
The planarity of the peptide bond is important for the stability and structure formation of proteins. However, substantial distortion of peptide bonds has been reported in several high-resolution structures and computational analyses. To investigate the peptide bond planarity, including hydrogen atoms, we report a 1.2-Å resolution neutron structure of the oxidized form of high-potential iron-sulfur protein. This high-resolution neutron structure shows that the nucleus positions of the amide protons deviate from the peptide plane and shift toward the acceptors. The planarity of the H─N─C═O plane depends strongly on the pyramidalization of the nitrogen atom. Moreover, the orientation of the amide proton of Cys75 is different in the reduced and oxidized states, possibly because of the electron storage capacity of the iron-sulfur cluster.
Figures
Similar articles
-
Site-specific relaxation of peptide bond planarity induced by electrically attracted proton/deuteron observed by neutron crystallography.Protein Sci. 2023 Oct;32(10):e4765. doi: 10.1002/pro.4765. Protein Sci. 2023. PMID: 37624071 Free PMC article.
-
Description of peptide bond planarity from high-resolution neutron crystallography.Biophys Physicobiol. 2023 Sep 6;20(3):e200035. doi: 10.2142/biophysico.bppb-v20.0035. eCollection 2023. Biophys Physicobiol. 2023. PMID: 38124796 Free PMC article.
-
Computational Study of Helical and Helix-Hinge-Helix Conformations of an Anti-Microbial Peptide in Solution by Molecular Dynamics and Vibrational Analysis.J Phys Chem B. 2021 Jan 28;125(3):703-721. doi: 10.1021/acs.jpcb.0c07988. Epub 2021 Jan 19. J Phys Chem B. 2021. PMID: 33464100
-
Evaluation of backbone proton positions and dynamics in a small protein by liquid crystal NMR spectroscopy.J Am Chem Soc. 2003 Jul 30;125(30):9179-91. doi: 10.1021/ja0350684. J Am Chem Soc. 2003. PMID: 15369375
-
Neutron Crystallography for the Study of Hydrogen Bonds in Macromolecules.Molecules. 2017 Apr 7;22(4):596. doi: 10.3390/molecules22040596. Molecules. 2017. PMID: 28387738 Free PMC article. Review.
Cited by
-
Site-specific relaxation of peptide bond planarity induced by electrically attracted proton/deuteron observed by neutron crystallography.Protein Sci. 2023 Oct;32(10):e4765. doi: 10.1002/pro.4765. Protein Sci. 2023. PMID: 37624071 Free PMC article.
-
Nitrogenase beyond the Resting State: A Structural Perspective.Molecules. 2023 Dec 5;28(24):7952. doi: 10.3390/molecules28247952. Molecules. 2023. PMID: 38138444 Free PMC article. Review.
-
Description of peptide bond planarity from high-resolution neutron crystallography.Biophys Physicobiol. 2023 Sep 6;20(3):e200035. doi: 10.2142/biophysico.bppb-v20.0035. eCollection 2023. Biophys Physicobiol. 2023. PMID: 38124796 Free PMC article.
References
-
- Smith R. M., Hansen D. E., The pH-rate profile for the hydrolysis of a peptide bond. J. Am. Chem. Soc. 120, 8910–8913 (1998).
-
- Bordo D., Argos P., The role of side-chain hydrogen bonds in the formation and stabilization of secondary structure in soluble proteins. J. Mol. Biol. 243, 504–519 (1994). - PubMed
-
- Eswar N., Ramakrishnan C., Secondary structures without backbone: An analysis of backbone mimicry by polar side chains in protein structures. Protein Eng. 12, 447–455 (1999). - PubMed

