Human M1 macrophages express unique innate immune response genes after mycobacterial infection to defend against tuberculosis
- PMID: 35590096
- PMCID: PMC9119986
- DOI: 10.1038/s42003-022-03387-9
Human M1 macrophages express unique innate immune response genes after mycobacterial infection to defend against tuberculosis
Abstract
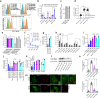
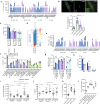
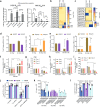
Mycobacterium tuberculosis (Mtb) is responsible for approximately 1.5 million deaths each year. Though 10% of patients develop tuberculosis (TB) after infection, 90% of these infections are latent. Further, mice are nearly uniformly susceptible to Mtb but their M1-polarized macrophages (M1-MΦs) can inhibit Mtb in vitro, suggesting that M1-MΦs may be able to regulate anti-TB immunity. We sought to determine whether human MΦ heterogeneity contributes to TB immunity. Here we show that IFN-γ-programmed M1-MΦs degrade Mtb through increased expression of innate immunity regulatory genes (Inregs). In contrast, IL-4-programmed M2-polarized MΦs (M2-MΦs) are permissive for Mtb proliferation and exhibit reduced Inregs expression. M1-MΦs and M2-MΦs express pro- and anti-inflammatory cytokine-chemokines, respectively, and M1-MΦs show nitric oxide and autophagy-dependent degradation of Mtb, leading to increased antigen presentation to T cells through an ATG-RAB7-cathepsin pathway. Despite Mtb infection, M1-MΦs show increased histone acetylation at the ATG5 promoter and pro-autophagy phenotypes, while increased histone deacetylases lead to decreased autophagy in M2-MΦs. Finally, Mtb-infected neonatal macaques express human Inregs in their lymph nodes and macrophages, suggesting that M1 and M2 phenotypes can mediate immunity to TB in both humans and macaques. We conclude that human MФ subsets show unique patterns of gene expression that enable differential control of TB after infection. These genes could serve as targets for diagnosis and immunotherapy of TB.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest and certify that all studies were conducted under approved institutional Health Science Center and Institutional review board protocols.
Figures




Similar articles
-
Mycobacterium tuberculosis Virulent Factor ESAT-6 Drives Macrophage Differentiation Toward the Pro-inflammatory M1 Phenotype and Subsequently Switches It to the Anti-inflammatory M2 Phenotype.Front Cell Infect Microbiol. 2018 Sep 18;8:327. doi: 10.3389/fcimb.2018.00327. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30283745 Free PMC article.
-
Polarization of Human Monocyte-Derived Cells With Vitamin D Promotes Control of Mycobacterium tuberculosis Infection.Front Immunol. 2020 Jan 22;10:3157. doi: 10.3389/fimmu.2019.03157. eCollection 2019. Front Immunol. 2020. PMID: 32038652 Free PMC article.
-
Natural and trained innate immunity against Mycobacterium tuberculosis.Immunobiology. 2020 May;225(3):151951. doi: 10.1016/j.imbio.2020.151951. Epub 2020 Apr 27. Immunobiology. 2020. PMID: 32423788 Review.
-
A CD4+CD161+ T-Cell Subset Present in Unexposed Humans, Not Tb Patients, Are Fast Acting Cells That Inhibit the Growth of Intracellular Mycobacteria Involving CD161 Pathway, Perforin, and IFN-γ/Autophagy.Front Immunol. 2021 Feb 26;12:599641. doi: 10.3389/fimmu.2021.599641. eCollection 2021. Front Immunol. 2021. PMID: 33732233 Free PMC article.
-
Functional Signatures of Human CD4 and CD8 T Cell Responses to Mycobacterium tuberculosis.Front Immunol. 2014 Apr 22;5:180. doi: 10.3389/fimmu.2014.00180. eCollection 2014. Front Immunol. 2014. PMID: 24795723 Free PMC article. Review.
Cited by
-
Triggering Receptor Expressed on Myeloid Cells 2 Mediates the Involvement of M2-Type Macrophages in Pulmonary Tuberculosis Infection.J Inflamm Res. 2024 Mar 27;17:1919-1928. doi: 10.2147/JIR.S435216. eCollection 2024. J Inflamm Res. 2024. PMID: 38562656 Free PMC article.
-
Marine-Fungi-Derived Gliotoxin Promotes Autophagy to Suppress Mycobacteria tuberculosis Infection in Macrophage.Mar Drugs. 2023 Nov 28;21(12):616. doi: 10.3390/md21120616. Mar Drugs. 2023. PMID: 38132937 Free PMC article.
-
Lung type 3 innate lymphoid cells respond early following Mycobacterium tuberculosis infection.mBio. 2024 Apr 10;15(4):e0329923. doi: 10.1128/mbio.03299-23. Epub 2024 Feb 26. mBio. 2024. PMID: 38407132 Free PMC article.
-
Exploring the Role of Extracellular Vesicles in the Pathogenesis of Tuberculosis.Genes (Basel). 2024 Mar 29;15(4):434. doi: 10.3390/genes15040434. Genes (Basel). 2024. PMID: 38674369 Free PMC article.
-
Identification and Validation of Genes Related to Macrophage Polarization and Cell Death Modes Under Mycobacterium tuberculosis Infection.J Inflamm Res. 2024 Feb 29;17:1397-1411. doi: 10.2147/JIR.S448372. eCollection 2024. J Inflamm Res. 2024. PMID: 38476473 Free PMC article.
References
-
- Koeken, V., Verrall, A. J., Netea, M. G., Hill, P. C. & van Crevel, R. Trained innate immunity and resistance to Mycobacterium tuberculosis infection. Clin. Microbiol. Infect.25, 1468–1472 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

