Lipid droplet degradation by autophagy connects mitochondria metabolism to Prox1-driven expression of lymphatic genes and lymphangiogenesis
- PMID: 35589749
- PMCID: PMC9120506
- DOI: 10.1038/s41467-022-30490-6
Lipid droplet degradation by autophagy connects mitochondria metabolism to Prox1-driven expression of lymphatic genes and lymphangiogenesis
Abstract
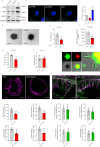
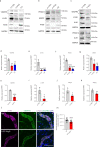
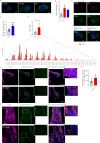
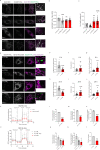
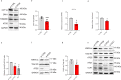
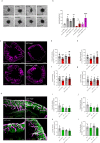
Autophagy has vasculoprotective roles, but whether and how it regulates lymphatic endothelial cells (LEC) homeostasis and lymphangiogenesis is unknown. Here, we show that genetic deficiency of autophagy in LEC impairs responses to VEGF-C and injury-driven corneal lymphangiogenesis. Autophagy loss in LEC compromises the expression of main effectors of LEC identity, like VEGFR3, affects mitochondrial dynamics and causes an accumulation of lipid droplets (LDs) in vitro and in vivo. When lipophagy is impaired, mitochondrial ATP production, fatty acid oxidation, acetyl-CoA/CoA ratio and expression of lymphangiogenic PROX1 target genes are dwindled. Enforcing mitochondria fusion by silencing dynamin-related-protein 1 (DRP1) in autophagy-deficient LEC fails to restore LDs turnover and lymphatic gene expression, whereas supplementing the fatty acid precursor acetate rescues VEGFR3 levels and signaling, and lymphangiogenesis in LEC-Atg5-/- mice. Our findings reveal that lipophagy in LEC by supporting FAO, preserves a mitochondrial-PROX1 gene expression circuit that safeguards LEC responsiveness to lymphangiogenic mediators and lymphangiogenesis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Eating your own fat to stay fit: lipophagy sustains lymphangiogenesis.Autophagy. 2023 Apr;19(4):1351-1353. doi: 10.1080/15548627.2022.2117513. Epub 2022 Sep 1. Autophagy. 2023. PMID: 36026459 Free PMC article.
-
The role of fatty acid β-oxidation in lymphangiogenesis.Nature. 2017 Feb 2;542(7639):49-54. doi: 10.1038/nature21028. Epub 2016 Dec 26. Nature. 2017. PMID: 28024299
-
YAP and TAZ Negatively Regulate Prox1 During Developmental and Pathologic Lymphangiogenesis.Circ Res. 2019 Jan 18;124(2):225-242. doi: 10.1161/CIRCRESAHA.118.313707. Circ Res. 2019. PMID: 30582452
-
Roles of prostaglandins in tumor-associated lymphangiogenesis with special reference to breast cancer.Cancer Metastasis Rev. 2018 Sep;37(2-3):369-384. doi: 10.1007/s10555-018-9734-0. Cancer Metastasis Rev. 2018. PMID: 29858743 Review.
-
Lymphatic endothelial cells, inflammatory lymphangiogenesis, and prospective players.Curr Med Chem. 2007;14(22):2359-68. doi: 10.2174/092986707781745541. Curr Med Chem. 2007. PMID: 17896984 Review.
Cited by
-
Lipid Droplet-Mitochondria Contacts in Health and Disease.Int J Mol Sci. 2024 Jun 22;25(13):6878. doi: 10.3390/ijms25136878. Int J Mol Sci. 2024. PMID: 38999988 Free PMC article. Review.
-
Exploring the roles and molecular mechanisms of RNA binding proteins in the sorting of noncoding RNAs into exosomes during tumor progression.J Adv Res. 2024 Nov;65:105-123. doi: 10.1016/j.jare.2023.11.029. Epub 2023 Nov 27. J Adv Res. 2024. PMID: 38030125 Free PMC article. Review.
-
Molecular probes for tracking lipid droplet membrane dynamics.Nat Commun. 2024 Oct 31;15(1):9413. doi: 10.1038/s41467-024-53667-7. Nat Commun. 2024. PMID: 39482302 Free PMC article.
-
Tumor endothelial cell autophagy is a key vascular-immune checkpoint in melanoma.EMBO Mol Med. 2023 Dec 7;15(12):e18028. doi: 10.15252/emmm.202318028. Epub 2023 Nov 27. EMBO Mol Med. 2023. PMID: 38009521 Free PMC article.
-
Decorin suppresses tumor lymphangiogenesis: A mechanism to curtail cancer progression.Proc Natl Acad Sci U S A. 2024 Apr 30;121(18):e2317760121. doi: 10.1073/pnas.2317760121. Epub 2024 Apr 23. Proc Natl Acad Sci U S A. 2024. PMID: 38652741 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

