A Test and Refinement of Folding Free Energy Nearest Neighbor Parameters for RNA Including N6-Methyladenosine
- PMID: 35588868
- PMCID: PMC11235186
- DOI: 10.1016/j.jmb.2022.167632
A Test and Refinement of Folding Free Energy Nearest Neighbor Parameters for RNA Including N6-Methyladenosine
Abstract
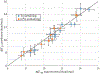
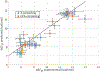
RNA folding free energy change parameters are widely used to predict RNA secondary structure and to design RNA sequences. These parameters include terms for the folding free energies of helices and loops. Although the full set of parameters has only been traditionally available for the four common bases and backbone, it is well known that covalent modifications of nucleotides are widespread in natural RNAs. Covalent modifications are also widely used in engineered sequences. We recently derived a full set of nearest neighbor terms for RNA that includes N6-methyladenosine (m6A). In this work, we test the model using 98 optical melting experiments, matching duplexes with or without N6-methylation of A. Most experiments place RRACH, the consensus site of N6-methylation, in a variety of contexts, including helices, bulge loops, internal loops, dangling ends, and terminal mismatches. For matched sets of experiments that include either A or m6A in the same context, we find that the parameters for m6A are as accurate as those for A. Across all experiments, the root mean squared deviation between estimated and experimental free energy changes is 0.67 kcal/mol. We used the new experimental data to refine the set of nearest neighbor parameter terms for m6A. These parameters enable prediction of RNA secondary structures including m6A, which can be used to model how N6-methylation of A affects RNA structure.
Keywords: RNA covalent modification; RNA folding stability; RNA methylation; RNA secondary structure prediction; optical melting.
Copyright © 2022 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests of personal relationships that could have appeared to influence the work reported in this paper.
Figures
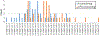

Similar articles
-
Analysis of RNA nearest neighbor parameters reveals interdependencies and quantifies the uncertainty in RNA secondary structure prediction.RNA. 2018 Nov;24(11):1568-1582. doi: 10.1261/rna.065102.117. Epub 2018 Aug 13. RNA. 2018. PMID: 30104207 Free PMC article.
-
Improving RNA nearest neighbor parameters for helices by going beyond the two-state model.Nucleic Acids Res. 2018 Jun 1;46(10):4883-4892. doi: 10.1093/nar/gky270. Nucleic Acids Res. 2018. PMID: 29718397 Free PMC article.
-
Secondary structure prediction for RNA sequences including N6-methyladenosine.Nat Commun. 2022 Mar 11;13(1):1271. doi: 10.1038/s41467-022-28817-4. Nat Commun. 2022. PMID: 35277476 Free PMC article.
-
The determination of RNA folding nearest neighbor parameters.Methods Mol Biol. 2014;1097:45-70. doi: 10.1007/978-1-62703-709-9_3. Methods Mol Biol. 2014. PMID: 24639154 Review.
-
The Functions of N6-Methyladenosine in Nuclear RNAs.Biochemistry (Mosc). 2024 Jan;89(1):159-172. doi: 10.1134/S0006297924010103. Biochemistry (Mosc). 2024. PMID: 38467552 Review.
Cited by
-
Tailor made: the art of therapeutic mRNA design.Nat Rev Drug Discov. 2024 Jan;23(1):67-83. doi: 10.1038/s41573-023-00827-x. Epub 2023 Nov 29. Nat Rev Drug Discov. 2024. PMID: 38030688 Review.
-
DNA Structure Design Is Improved Using an Artificially Expanded Alphabet of Base Pairs Including Loop and Mismatch Thermodynamic Parameters.bioRxiv [Preprint]. 2023 Jun 8:2023.06.06.543917. doi: 10.1101/2023.06.06.543917. bioRxiv. 2023. Update in: ACS Synth Biol. 2023 Sep 15;12(9):2750-2763. doi: 10.1021/acssynbio.3c00358 PMID: 37333404 Free PMC article. Updated. Preprint.
-
m6A readers, writers, erasers, and the m6A epitranscriptome in breast cancer.J Mol Endocrinol. 2022 Dec 21;70(2):e220110. doi: 10.1530/JME-22-0110. Print 2023 Feb 1. J Mol Endocrinol. 2022. PMID: 36367225 Free PMC article. Review.
-
Molecular insight into how the position of an abasic site modifies DNA duplex stability and dynamics.Biophys J. 2024 Jan 16;123(2):118-133. doi: 10.1016/j.bpj.2023.11.022. Epub 2023 Nov 24. Biophys J. 2024. PMID: 38006207
-
RNA Secondary Structure Analysis Using RNAstructure.Curr Protoc. 2023 Jul;3(7):e846. doi: 10.1002/cpz1.846. Curr Protoc. 2023. PMID: 37487054 Free PMC article.
References
-
- Li X, Xiong X, Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods. 2016;14:23–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

