Temporal relationship among adiposity, gut microbiota, and insulin resistance in a longitudinal human cohort
- PMID: 35585555
- PMCID: PMC9118787
- DOI: 10.1186/s12916-022-02376-3
Temporal relationship among adiposity, gut microbiota, and insulin resistance in a longitudinal human cohort
Abstract
Background: The temporal relationship between adiposity and gut microbiota was unexplored. Whether some gut microbes lie in the pathways from adiposity to insulin resistance is less clear. Our study aims to reveal the temporal relationship between adiposity and gut microbiota and investigate whether gut microbiota may mediate the association of adiposity with insulin resistance in a longitudinal human cohort study.
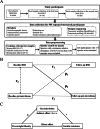
Methods: We obtained repeated-measured gut shotgun metagenomic and anthropometric data from 426 Chinese participants over ~3 years of follow-up. Cross-lagged path analysis was used to examine the temporal relationship between BMI and gut microbial features. The associations between the gut microbes and insulin resistance-related phenotypes were examined using a linear mixed-effect model. We examined the mediation effect of gut microbes on the association between adiposity and insulin resistance-related phenotypes. Replication was performed in the HMP cohort.
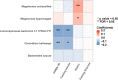
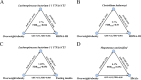
Results: Baseline BMI was prospectively associated with levels of ten gut microbial species. Among them, results of four species (Adlercreutzia equolifaciens, Parabacteroides unclassified, Lachnospiraceae bacterium 3 1 57FAA CT1, Lachnospiraceae bacterium 7 1 58FAA) were replicated in the independent HMP cohort. Lachnospiraceae bacterium 3 1 57FAA CT1 was inversely associated with HOMA-IR and fasting insulin. Lachnospiraceae bacterium 3 1 57FAA CT1 mediated the association of overweight/obesity with HOMA-IR (FDR<0.05). Furthermore, Lachnospiraceae bacterium 3 1 57FAA CT1 was positively associated with the butyrate-producing pathway PWY-5022 (p < 0.001).
Conclusions: Our study identified one potentially beneficial microbe Lachnospiraceae bacterium 3 1 57FAA CT1, which might mediate the effect of adiposity on insulin resistance. The identified microbes are helpful for the discovery of novel therapeutic targets, as to mitigate the impact of adiposity on insulin resistance.
Keywords: Adiposity; Gut microbiota; Insulin resistance; Longitudinal cohort study; Obesity; Temporal relationship; Weight change.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Metagenomic Shotgun Sequencing Reveals Specific Human Gut Microbiota Associated with Insulin Resistance and Body Fat Distribution in Saudi Women.Biomolecules. 2023 Apr 2;13(4):640. doi: 10.3390/biom13040640. Biomolecules. 2023. PMID: 37189387 Free PMC article.
-
Association of gut microbiota with glycaemic traits and incident type 2 diabetes, and modulation by habitual diet: a population-based longitudinal cohort study in Chinese adults.Diabetologia. 2022 Jul;65(7):1145-1156. doi: 10.1007/s00125-022-05687-5. Epub 2022 Mar 31. Diabetologia. 2022. PMID: 35357559 Free PMC article.
-
Gut microbiota and acylcarnitine metabolites connect the beneficial association between equol and adiposity in adults: a prospective cohort study.Am J Clin Nutr. 2022 Dec 19;116(6):1831-1841. doi: 10.1093/ajcn/nqac252. Am J Clin Nutr. 2022. PMID: 36095141 Clinical Trial.
-
Obesity and the human microbiome.Curr Opin Gastroenterol. 2010 Jan;26(1):5-11. doi: 10.1097/MOG.0b013e328333d751. Curr Opin Gastroenterol. 2010. PMID: 19901833 Review.
-
Gut microbiome and its role in obesity and insulin resistance.Ann N Y Acad Sci. 2020 Feb;1461(1):37-52. doi: 10.1111/nyas.14107. Epub 2019 May 14. Ann N Y Acad Sci. 2020. PMID: 31087391 Review.
Cited by
-
Temporal relationship between hepatic steatosis and blood pressure elevation and the mediation effect in the development of cardiovascular disease.Hypertens Res. 2024 Jul;47(7):1811-1821. doi: 10.1038/s41440-024-01708-5. Epub 2024 May 17. Hypertens Res. 2024. PMID: 38760520
-
Longitudinal profiling of the microbiome at four body sites reveals core stability and individualized dynamics during health and disease.Cell Host Microbe. 2024 Apr 10;32(4):506-526.e9. doi: 10.1016/j.chom.2024.02.012. Epub 2024 Mar 12. Cell Host Microbe. 2024. PMID: 38479397
-
Effects of vitamin D supplementation on glucose and lipid metabolism in patients with type 2 diabetes mellitus and risk factors for insulin resistance.World J Diabetes. 2023 Oct 15;14(10):1514-1523. doi: 10.4239/wjd.v14.i10.1514. World J Diabetes. 2023. PMID: 37970127 Free PMC article.
-
The association between BMI and serum uric acid is partially mediated by gut microbiota.Microbiol Spectr. 2023 Sep 25;11(5):e0114023. doi: 10.1128/spectrum.01140-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37747198 Free PMC article.
-
Diet and the Gut Microbiome as Determinants Modulating Metabolic Outcomes in Young Obese Adults.Biomedicines. 2024 Jul 18;12(7):1601. doi: 10.3390/biomedicines12071601. Biomedicines. 2024. PMID: 39062174 Free PMC article.
References
-
- Obesity and overweight. https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight. Accessed 11 Oct 2021.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

