Regulation of neutrophil myeloperoxidase inhibitor SPIN by the small RNA Teg49 in Staphylococcus aureus
- PMID: 35578788
- PMCID: PMC9880452
- DOI: 10.1111/mmi.14919
Regulation of neutrophil myeloperoxidase inhibitor SPIN by the small RNA Teg49 in Staphylococcus aureus
Abstract
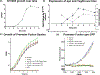
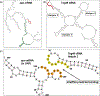
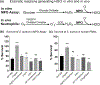
Teg49 is a Staphylococcus aureus trans-acting regulatory sRNA derived from cleavage of the sarA P3 transcript. We showed by RNA-Seq here that the 5' trident-like structure in Teg49 regulates transcriptionally (direct and indirect) 22 genes distinct from sarA. Among these, Teg49 was noted to repress spn, encoding a 102 residue preprotein which yields the mature 73 residue peptide which inhibits the catalytic activity of myeloperoxidase in human neutrophils. Teg49 was found to regulate spn mRNA post-transcriptionally in strain SH1000 through 9-nt base-pairing between hairpin loop 2 of Teg49 and an exposed bulge of the spn mRNA. Mutations of the Teg49 binding site disrupted the repression of spn, leading to reduced degradation, and increased half-life of spn mRNA in the Teg49 mutant. The spn-Teg49 interaction was also confirmed with a synonymous spn mutation to yield enhanced spn expression in the mutant vs. the parent. The Teg49 mutant with increased spn expression exhibited enhanced resistance to MPO activity in vitro. Killing assays with human neutrophils showed that the Teg49 mutant was more resistant to killing after phagocytosis. Altogether, this study shows that Teg49 in S. aureus has a distinct and important regulatory profile whereby this sRNA modulates resistance to myeloperoxidase-mediated killing by human neutrophils.
Keywords: Staphylococcus aureus; spn; RNA; SPIN; gene expression; human neutrophils; myeloperoxidase; regulatory small RNA; virulence.
© 2022 John Wiley & Sons Ltd.
Figures
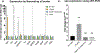

Similar articles
-
Small RNA teg49 Is Derived from a sarA Transcript and Regulates Virulence Genes Independent of SarA in Staphylococcus aureus.Infect Immun. 2018 Jan 22;86(2):e00635-17. doi: 10.1128/IAI.00635-17. Print 2018 Feb. Infect Immun. 2018. PMID: 29133345 Free PMC article.
-
Contribution of teg49 small RNA in the 5' upstream transcriptional region of sarA to virulence in Staphylococcus aureus.Infect Immun. 2014 Oct;82(10):4369-79. doi: 10.1128/IAI.02002-14. Epub 2014 Aug 4. Infect Immun. 2014. PMID: 25092913 Free PMC article.
-
Antibacterial activity of silver nanoparticles target sara through srna-teg49, a key mediator of hfq, in staphylococcus aureus.Int J Clin Exp Med. 2015 Apr 15;8(4):5794-9. eCollection 2015. Int J Clin Exp Med. 2015. PMID: 26131167 Free PMC article.
-
The power of cooperation: Experimental and computational approaches in the functional characterization of bacterial sRNAs.Mol Microbiol. 2020 Mar;113(3):603-612. doi: 10.1111/mmi.14420. Epub 2019 Nov 28. Mol Microbiol. 2020. PMID: 31705780 Free PMC article. Review.
-
Mechanistic study of base-pairing small regulatory RNAs in bacteria.Methods. 2017 Mar 15;117:67-76. doi: 10.1016/j.ymeth.2016.09.012. Epub 2016 Sep 28. Methods. 2017. PMID: 27693881 Review.
Cited by
-
Thirty Years of sRNA-Mediated Regulation in Staphylococcus aureus: From Initial Discoveries to In Vivo Biological Implications.Int J Mol Sci. 2022 Jul 1;23(13):7346. doi: 10.3390/ijms23137346. Int J Mol Sci. 2022. PMID: 35806357 Free PMC article. Review.
References
-
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W et al. (2013) Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nature Protocols, 8, 1765–1786. - PubMed
-
- Andronescu M, Condon A, Hoos HH, Mathews DH & Murphy KP (2007) Efficient parameter estimation for RNA secondary structure prediction. Bioinformatics, 23, i19–i28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

