Epigenetic and transcriptional dysregulation in CD4+ T cells in patients with atopic dermatitis
- PMID: 35576187
- PMCID: PMC9135339
- DOI: 10.1371/journal.pgen.1009973
Epigenetic and transcriptional dysregulation in CD4+ T cells in patients with atopic dermatitis
Abstract
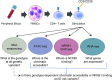
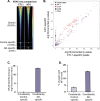
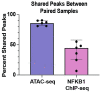
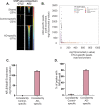
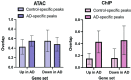
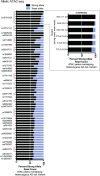
Atopic dermatitis (AD) is one of the most common skin disorders among children. Disease etiology involves genetic and environmental factors, with 29 independent AD risk loci enriched for risk allele-dependent gene expression in the skin and CD4+ T cell compartments. We investigated the potential epigenetic mechanisms responsible for the genetic susceptibility of CD4+ T cells. To understand the differences in gene regulatory activity in peripheral blood T cells in AD, we measured chromatin accessibility (an assay based on transposase-accessible chromatin sequencing, ATAC-seq), nuclear factor kappa B subunit 1 (NFKB1) binding (chromatin immunoprecipitation with sequencing, ChIP-seq), and gene expression levels (RNA-seq) in stimulated CD4+ T cells from subjects with active moderate-to-severe AD, as well as in age-matched non-allergic controls. Open chromatin regions in stimulated CD4+ T cells were highly enriched for AD genetic risk variants, with almost half of the AD risk loci overlapping AD-dependent ATAC-seq peaks. AD-specific open chromatin regions were strongly enriched for NF-κB DNA-binding motifs. ChIP-seq identified hundreds of NFKB1-occupied genomic loci that were AD- or control-specific. As expected, the AD-specific ChIP-seq peaks were strongly enriched for NF-κB DNA-binding motifs. Surprisingly, control-specific NFKB1 ChIP-seq peaks were not enriched for NFKB1 motifs, but instead contained motifs for other classes of human transcription factors, suggesting a mechanism involving altered indirect NFKB1 binding. Using DNA sequencing data, we identified 63 instances of altered genotype-dependent chromatin accessibility at 36 AD risk variant loci (30% of AD risk loci) that might lead to genotype-dependent gene expression. Based on these findings, we propose that CD4+ T cells respond to stimulation in an AD-specific manner, resulting in disease- and genotype-dependent chromatin accessibility alterations involving NFKB1 binding.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Integrated analysis of ATAC-seq and RNA-seq reveals the transcriptional regulation network in SLE.Int Immunopharmacol. 2023 Mar;116:109803. doi: 10.1016/j.intimp.2023.109803. Epub 2023 Feb 2. Int Immunopharmacol. 2023. PMID: 36738683
-
Epigenetic Landscapes of Single-Cell Chromatin Accessibility and Transcriptomic Immune Profiles of T Cells in COVID-19 Patients.Front Immunol. 2021 Feb 24;12:625881. doi: 10.3389/fimmu.2021.625881. eCollection 2021. Front Immunol. 2021. PMID: 33717140 Free PMC article.
-
Epigenetic dysregulation in Alzheimer's disease peripheral immunity.Neuron. 2024 Apr 17;112(8):1235-1248.e5. doi: 10.1016/j.neuron.2024.01.013. Epub 2024 Feb 9. Neuron. 2024. PMID: 38340719
-
[Advances in assay for transposase-accessible chromatin with high-throughput sequencing].Yi Chuan. 2020 Apr 20;42(4):333-346. doi: 10.16288/j.yczz.19-279. Yi Chuan. 2020. PMID: 32312702 Review. Chinese.
-
Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond.Cell Cycle. 2014;13(18):2847-52. doi: 10.4161/15384101.2014.949201. Cell Cycle. 2014. PMID: 25486472 Free PMC article. Review.
Cited by
-
Rapid profiling of transcription factor-cofactor interaction networks reveals principles of epigenetic regulation.bioRxiv [Preprint]. 2024 Apr 6:2024.04.05.588333. doi: 10.1101/2024.04.05.588333. bioRxiv. 2024. Update in: Nucleic Acids Res. 2024 Sep 23;52(17):10276-10296. doi: 10.1093/nar/gkae706. PMID: 38617258 Free PMC article. Updated. Preprint.
-
Rapid profiling of transcription factor-cofactor interaction networks reveals principles of epigenetic regulation.Nucleic Acids Res. 2024 Sep 23;52(17):10276-10296. doi: 10.1093/nar/gkae706. Nucleic Acids Res. 2024. PMID: 39166482 Free PMC article.
-
maxATAC: Genome-scale transcription-factor binding prediction from ATAC-seq with deep neural networks.PLoS Comput Biol. 2023 Jan 31;19(1):e1010863. doi: 10.1371/journal.pcbi.1010863. eCollection 2023 Jan. PLoS Comput Biol. 2023. PMID: 36719906 Free PMC article.
-
Molecular characterization of allergic constitution based on network pharmacology and multi-omics analysis methods.Medicine (Baltimore). 2024 Feb 16;103(7):e36892. doi: 10.1097/MD.0000000000036892. Medicine (Baltimore). 2024. PMID: 38363941 Free PMC article.
-
Omics and Lung Function: A Need for Integration.Am J Respir Crit Care Med. 2022 Aug 1;206(3):242-244. doi: 10.1164/rccm.202205-0928ED. Am J Respir Crit Care Med. 2022. PMID: 35608542 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- U01 AI130830/AI/NIAID NIH HHS/United States
- R01 NS099068/NS/NINDS NIH HHS/United States
- KL2 TR001426/TR/NCATS NIH HHS/United States
- R01 DK107502/DK/NIDDK NIH HHS/United States
- R01 HG010730/HG/NHGRI NIH HHS/United States
- R01 GM055479/GM/NIGMS NIH HHS/United States
- R01 AI024717/AI/NIAID NIH HHS/United States
- P30 AR070549/AR/NIAMS NIH HHS/United States
- R01 AI148276/AI/NIAID NIH HHS/United States
- U19 AI070235/AI/NIAID NIH HHS/United States
- R01 AR073228/AR/NIAMS NIH HHS/United States
- U01 HG011172/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

