In Silico Analysis Predicts a Limited Impact of SARS-CoV-2 Variants on CD8 T Cell Recognition
- PMID: 35572563
- PMCID: PMC9094405
- DOI: 10.3389/fimmu.2022.891524
In Silico Analysis Predicts a Limited Impact of SARS-CoV-2 Variants on CD8 T Cell Recognition
Abstract
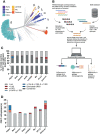
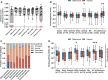
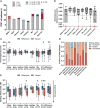
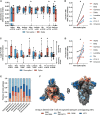
Since the start of the COVID-19 pandemic, mutations have led to the emergence of new SARS-CoV-2 variants, and some of these have become prominent or dominant variants of concern. This natural course of development can have an impact on how protective the previously naturally or vaccine induced immunity is. Therefore, it is crucial to understand whether and how variant specific mutations influence host immunity. To address this, we have investigated how mutations in the recent SARS-CoV-2 variants of interest and concern influence epitope sequence similarity, predicted binding affinity to HLA, and immunogenicity of previously reported SARS-CoV-2 CD8 T cell epitopes. Our data suggests that the vast majority of SARS-CoV-2 CD8 T cell recognized epitopes are not altered by variant specific mutations. Interestingly, for the CD8 T cell epitopes that are altered due to variant specific mutations, our analyses show there is a high degree of sequence similarity between mutated and reference SARS-CoV-2 CD8 T cell epitopes. However, mutated epitopes, primarily derived from the spike protein, in SARS-CoV-2 variants Delta, AY.4.2 and Mu display reduced predicted binding affinity to their restriction element. These findings indicate that the recent SARS-CoV-2 variants of interest and concern have limited ability to escape memory CD8 T cell responses raised by vaccination or prior infection with SARS-CoV-2 early in the pandemic. The overall low impact of the mutations on CD8 T cell cross-recognition is in accordance with the notion that mutations in SARS-CoV-2 are primarily the result of receptor binding affinity and antibody selection pressures exerted on the spike protein, unrelated to T cell immunity.
Keywords: CD8 T cell epitopes; CD8 T cells; SARS-CoV-2; SARS-CoV-2 variants; bioinformatics & computational biology.
Copyright © 2022 Isaeva, Ketelaars and Kvistborg.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
CD8+ T-Cell Epitope Variations Suggest a Potential Antigen HLA-A2 Binding Deficiency for Spike Protein of SARS-CoV-2.Front Immunol. 2022 Jan 18;12:764949. doi: 10.3389/fimmu.2021.764949. eCollection 2021. Front Immunol. 2022. PMID: 35116022 Free PMC article.
-
A bioinformatic analysis of T-cell epitope diversity in SARS-CoV-2 variants: association with COVID-19 clinical severity in the United States population.Front Immunol. 2024 May 9;15:1357731. doi: 10.3389/fimmu.2024.1357731. eCollection 2024. Front Immunol. 2024. PMID: 38784379 Free PMC article.
-
In silico analysis of mutant epitopes in new SARS-CoV-2 lineages suggest global enhanced CD8+ T cell reactivity and also signs of immune response escape.Infect Genet Evol. 2022 Apr;99:105236. doi: 10.1016/j.meegid.2022.105236. Epub 2022 Feb 8. Infect Genet Evol. 2022. PMID: 35149224 Free PMC article.
-
Degenerate CD8 Epitopes Mapping to Structurally Constrained Regions of the Spike Protein: A T Cell-Based Way-Out From the SARS-CoV-2 Variants Storm.Front Immunol. 2021 Sep 8;12:730051. doi: 10.3389/fimmu.2021.730051. eCollection 2021. Front Immunol. 2021. PMID: 34566990 Free PMC article. Review.
-
Evolution of SARS-CoV-2-specific CD4+ T cell epitopes.Immunogenetics. 2023 Jun;75(3):283-293. doi: 10.1007/s00251-023-01295-8. Epub 2023 Jan 31. Immunogenetics. 2023. PMID: 36719467 Free PMC article. Review.
References
-
- Lam ME. United by the Global COVID-19 Pandemic: Divided by Our Values and Viral Identities. Humanities Soc Sci Commun (2021) 8:1–6. doi: 10.1057/s41599-020-00679-5 - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

