A bioinformatic analysis of WFDC2 (HE4) expression in high grade serous ovarian cancer reveals tumor-specific changes in metabolic and extracellular matrix gene expression
- PMID: 35568777
- PMCID: PMC9107348
- DOI: 10.1007/s12032-022-01665-4
A bioinformatic analysis of WFDC2 (HE4) expression in high grade serous ovarian cancer reveals tumor-specific changes in metabolic and extracellular matrix gene expression
Abstract
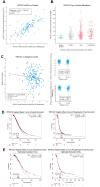
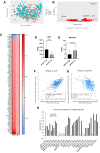
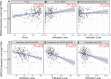
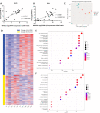
Human epididymis protein-4 (HE4/WFDC2) has been well-studied as an ovarian cancer clinical biomarker. To improve our understanding of its functional role in high grade serous ovarian cancer, we determined transcriptomic differences between ovarian tumors with high- versus low-WFDC2 mRNA levels in The Cancer Genome Atlas dataset. High-WFDC2 transcript levels were significantly associated with reduced survival in stage III/IV serous ovarian cancer patients. Differential expression and correlation analyses revealed secretory leukocyte peptidase inhibitor (SLPI/WFDC4) as the gene most positively correlated with WFDC2, while A kinase anchor protein-12 was most negatively correlated. WFDC2 and SLPI were strongly correlated across many cancers. Gene ontology analysis revealed enrichment of oxidative phosphorylation in differentially expressed genes associated with high-WFDC2 levels, while extracellular matrix organization was enriched among genes associated with low-WFDC2 levels. Immune cell subsets found to be positively correlated with WFDC2 levels were B cells and plasmacytoid dendritic cells, while neutrophils and endothelial cells were negatively correlated with WFDC2. Results were compared with DepMap cell culture gene expression data. Gene ontology analysis of k-means clustering revealed that genes associated with low-WFDC2 were also enriched in extracellular matrix and adhesion categories, while high-WFDC2 genes were enriched in epithelial cell proliferation and peptidase activity. These results support previous findings regarding the effect of HE4/WFDC2 on ovarian cancer pathogenesis in cell lines and mouse models, while adding another layer of complexity to its potential functions in ovarian tumor tissue. Further experimental explorations of these findings in the context of the tumor microenvironment are merited.
Keywords: HE4; Ovarian cancer; SLPI; The Cancer Genome Atlas; Tumor microenvironment; WFDC2.
© 2022. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
The biomarker HE4 (WFDC2) promotes a pro-angiogenic and immunosuppressive tumor microenvironment via regulation of STAT3 target genes.Sci Rep. 2020 May 22;10(1):8558. doi: 10.1038/s41598-020-65353-x. Sci Rep. 2020. PMID: 32444701 Free PMC article.
-
[Analysis of the correlation between the expression of WFDC2 in B cells of lung adenocarcinoma tissue and the survival rate of patients].Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2019 May;35(5):425-433. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2019. PMID: 31223112 Chinese.
-
WFDC2 (HE4): a potential role in the innate immunity of the oral cavity and respiratory tract and the development of adenocarcinomas of the lung.Respir Res. 2006 Apr 6;7(1):61. doi: 10.1186/1465-9921-7-61. Respir Res. 2006. PMID: 16600032 Free PMC article.
-
Ovarian cancer and normal fallopian tube high WFDC2 expression does not correlate with HE4 serum level.Ginekol Pol. 2015 May;86(5):335-9. doi: 10.17772/gp/2418. Ginekol Pol. 2015. PMID: 26117969
-
The emerging role of HE4 in the evaluation of epithelial ovarian and endometrial carcinomas.Oncology (Williston Park). 2013 Jun;27(6):548-56. Oncology (Williston Park). 2013. PMID: 23909069 Free PMC article. Review.
Cited by
-
HE4-based nomogram for predicting overall survival in patients with idiopathic pulmonary fibrosis: construction and validation.Eur J Med Res. 2024 Apr 16;29(1):238. doi: 10.1186/s40001-024-01829-0. Eur J Med Res. 2024. PMID: 38627872 Free PMC article.
-
Exploring the cellular and molecular differences between ovarian clear cell carcinoma and high-grade serous carcinoma using single-cell RNA sequencing and GEO gene expression signatures.Cell Biosci. 2023 Jul 31;13(1):139. doi: 10.1186/s13578-023-01087-3. Cell Biosci. 2023. PMID: 37525249 Free PMC article.
References
-
- Hellström I, et al. The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer Res. 2003;63:3695–3700. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

